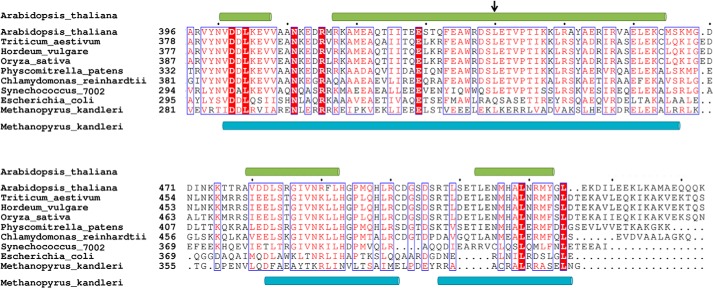
FIGURE 5.
Multiple sequence alignment of the C-terminal regions of GluTRs. Identical amino acid residues are boxed in red, and similar residues are printed in red in a blue box. Dots indicate gaps introduced during alignment. The secondary structure elements, as observed in the GluTR structures from Arabidopsis thaliana and Methanopyrus kandleri, are shown on the top and the bottom of the aligned sequences, respectively. The N-terminal end of GluTRDD in the FLUTPR-GluTRDD structure is indicated by an arrow.