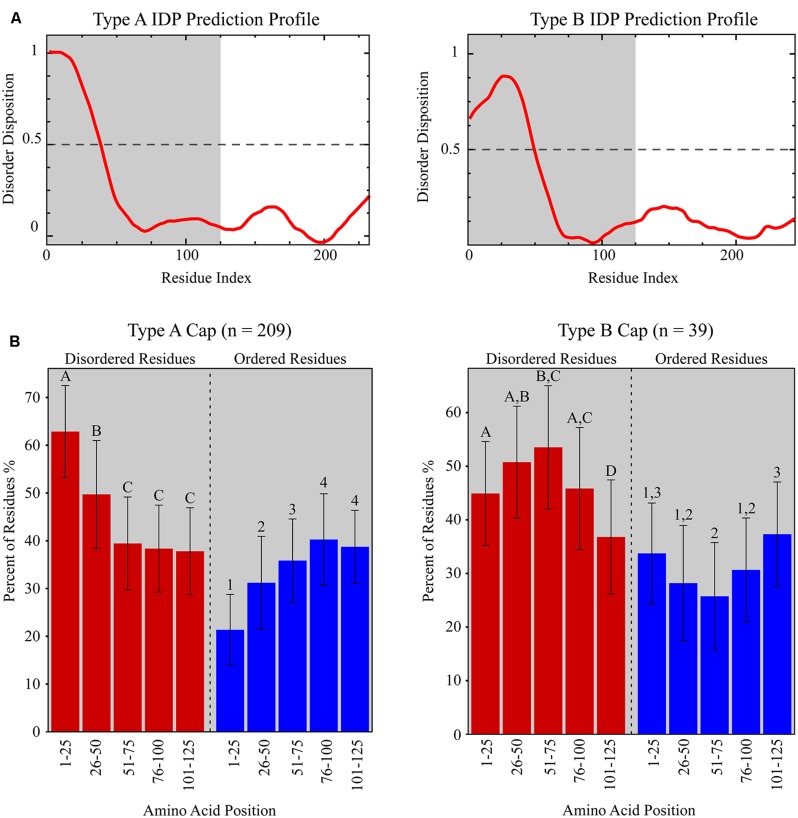
FIGURE 4.
(A) Representative IDP prediction profiles for Type A and Type B capsid proteins (Caps) from the Disprot VL3 predictor. Type A and Type B IDP prediction profiles are based on the Porcine circovirus 2 Cap (NP_937957.1) and the Beak and feather disease virus Cap (NP_047277.1), respectively. The grey shaded area represents the amino acid residue interval used in (B). (B) Graphs showing the fraction of predicted disordered (red bars) and ordered (blue bars) residues within discrete amino acid intervals for Type A and Type B Caps identified from all CRESS-DNA viral genomes analyzed in this study. Significantly different amino acid intervals for each Cap type are distinguished using letters (“A”, “B”, “C”, “D” for statistics based on percentage of predicted disordered residues) or numbers (“1”, “2”, “3”, “4” for statistics based on percentage of predicted ordered residues; ANOVA with post hoc Tukey’s HSD; p < 0.05). Note that the percentage of predicted disordered and ordered residues does not add to 100% due to the presence of residues that are not considered either disordered or ordered (i.e., H, M, T, and D).