MYC5 is a target of JAZs and functions redundantly with MYC2, MYC3, and MYC4 to regulate stamen development and seed production via interactions with the MYB transcription factors MYB21 and MYB24.
Abstract
Stamens are the plant male reproductive organs essential for plant fertility. Proper development of stamens is modulated by environmental cues and endogenous hormone signals. Deficiencies in biosynthesis or perception of the phytohormone jasmonate (JA) attenuate stamen development, disrupt male fertility, and abolish seed production in Arabidopsis thaliana. This study revealed that JA-mediated stamen development and seed production are regulated by a bHLH-MYB complex. The IIIe basic helix-loop-helix (bHLH) transcription factor MYC5 acts as a target of JAZ repressors to function redundantly with other IIIe bHLH factors such as MYC2, MYC3, and MYC4 in the regulation of stamen development and seed production. The myc2 myc3 myc4 myc5 quadruple mutant exhibits obvious defects in stamen development and significant reduction in seed production. Moreover, these IIIe bHLH factors interact with the MYB transcription factors MYB21 and MYB24 to form a bHLH-MYB transcription complex and cooperatively regulate stamen development. We speculate that the JAZ proteins repress the bHLH-MYB complex to suppress stamen development and seed production, while JA induces JAZ degradation and releases the bHLH-MYB complex to subsequently activate the expression of downstream genes essential for stamen development and seed production.
INTRODUCTION
As the male reproductive organ in plants, stamens are essential for fertility, and defects in stamen development can result in male sterility (Sanders et al., 1999). The development of stamens, comprising filament elongation, anther dehiscence, and pollen maturation at various floral developmental stages (Regan and Moffatt, 1990; Smyth et al., 1990), is regulated by diverse environmental cues and endogenous hormone signals, including gibberellin and jasmonate (JA) (Ma, 2005; Plackett et al., 2011; Song et al., 2013b).
JAs are a class of lipid-derived phytohormone (Browse, 2009; Wasternack and Hause, 2013) that regulate various essential plant defense responses and diverse developmental processes (Howe and Jander, 2008; Sun et al., 2011; De Geyter et al., 2012; Farmer and Mueller, 2013; Xin and He, 2013; Kazan and Lyons, 2014; Song et al., 2014b). Deficiencies in the biosynthesis or perception of JA attenuate stamen development, disrupt male fertility, and abolish seed production in various plant species, including both monocot (Acosta et al., 2009; Yan et al., 2012; Cai et al., 2014) and dicot plants (Feys et al., 1994; McConn and Browse, 1996). Specifically, Arabidopsis thaliana mutants deficient in JA perception, signaling, and biosynthesis, such as coronatine insensitive1 (coi1), JAZ1Δ3A, fad3 fad7 fad8, defective in anther dehiscence1, allene oxide synthase, and oxophytodienoate reductase3 (opr3), are defective in filament elongation, anther dehiscence, and pollen maturation at floral stage 13 and unable to produce seeds (Feys et al., 1994; McConn and Browse, 1996; Sanders et al., 2000; Stintzi and Browse, 2000; Ishiguro et al., 2001; Park et al., 2002; von Malek et al., 2002; Thines et al., 2007; Chung et al., 2010; Huang et al., 2014).
The Arabidopsis R2R3 MYB transcription factors MYB21, MYB24, and MYB57 (Mandaokar et al., 2006; Yang et al., 2007; Cheng et al., 2009) function redundantly to regulate stamen development. Another R2R3 MYB factor, MYB108, may act downstream in the regulation of anther dehiscence (Mandaokar and Browse, 2009). JA induces the expression of MYB21, MYB24, MYB57, and MYB108 (Mandaokar et al., 2006), while the JAZ repressors interact with and repress MYB21, MYB24, and MYB57 to modulate JA-regulated stamen development (Song et al., 2011).
The JAZ repressors regulate various transcription factors to influence diverse JA responses (Cheng et al., 2011; Niu et al., 2011; Pauwels and Goossens, 2011; Qi et al., 2011; Song et al., 2011, 2013a; Zhu et al., 2011; Kazan and Manners, 2012; Fonseca et al., 2014). The subgroup IIIe basic helix-loop-helix (bHLH) transcription factors MYC2, MYC3, and MYC4, which are direct targets of JAZ proteins (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011), form homodimers/heterodimers and function redundantly to regulate JA-mediated root growth inhibition, defense against insects, resistance to pathogen infection, and secondary metabolism (Boter et al., 2004; Lorenzo et al., 2004; Yadav et al., 2005; Dombrecht et al., 2007; Chen et al., 2011; Fernández-Calvo et al., 2011; Hong et al., 2012; Schweizer et al., 2013; Chico et al., 2014).
Here, we found that the IIIe bHLH transcription factor MYC5 is a target of JAZ repressors and functions redundantly with the other IIIe bHLH factors MYC2, MYC3, and MYC4 in the regulation of stamen development and seed production. Moreover, these IIIe bHLH factors interact with the R2R3 MYB transcription factors MYB21 and MYB24 to form a bHLH-MYB transcription complex and cooperatively regulate stamen development. Our results thus identify a bHLH-MYB complex responsible for the regulation of JA-mediated stamen development and seed production.
RESULTS
JAZ Proteins Interact with MYC5
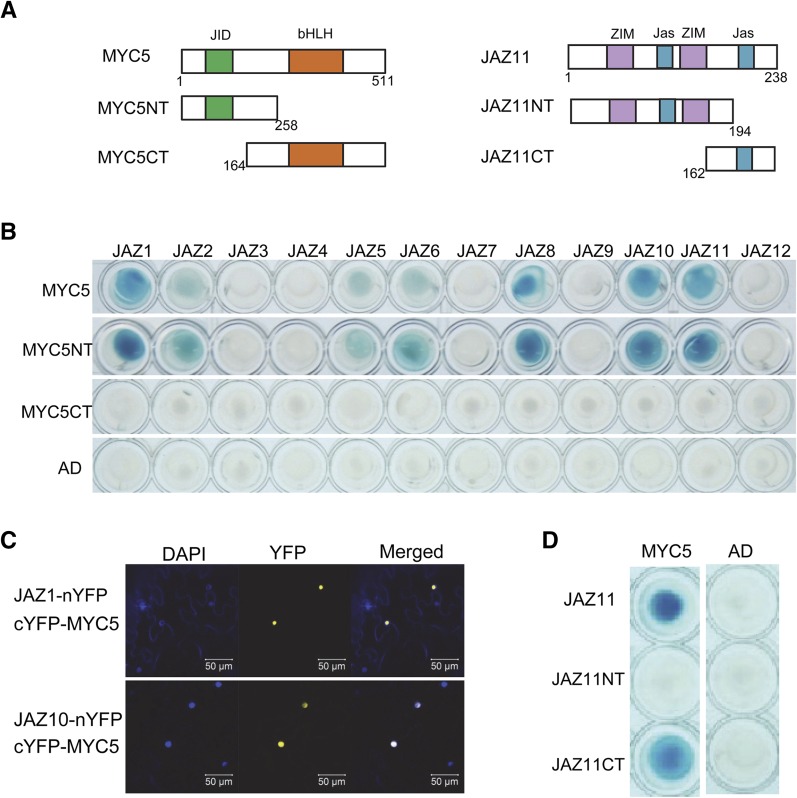
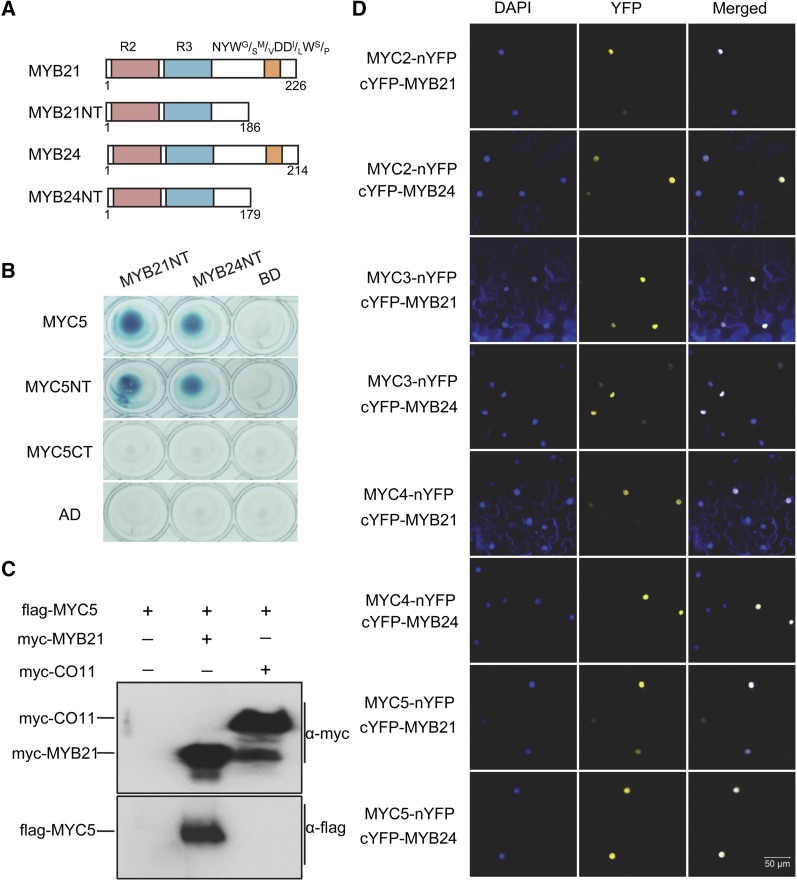
We exhaustively screened an Arabidopsis cDNA library using JAZ1 protein as bait in the yeast two-hybrid (Y2H) system (Qi et al., 2011; Song et al., 2011, 2013a) and found that a bHLH transcription factor, MYC5, interacted with JAZ proteins (Figures 1A and 1B). As shown in Figure 1B, Y2H analysis showed that MYC5 exhibited interaction with JAZ1, JAZ2, JAZ5, JAZ6, JAZ8, JAZ10, and JAZ11 in yeast. Yellow fluorescent protein (YFP)-based bimolecular fluorescence complementation (BiFC) assays showed that coexpression of JAZ1-nYFP (JAZ1 fusion with the N-terminal part of YFP) or JAZ10-nYFP with cYFP-MYC5 (MYC5 fusion with the C-terminal part of YFP) reconstructed strong YFP signals in the nuclei (Figure 1C; Supplemental Figure 1), demonstrating the interaction of JAZ1 and JAZ10 with MYC5 in planta. Further domain analysis showed that the N terminus of MYC5 containing the JAZ-interaction domain (Fernández-Calvo et al., 2011) is critical for interaction with JAZ proteins (Figures 1A and 1B), whereas the C-terminal part of JAZ11 harboring the Jas domain is necessary for JAZ interaction with MYC5 (Figures 1A and 1D). Taken together (Figure 1), these Y2H and BiFC results demonstrate that MYC5 is a target of JAZ proteins.
Figure 1.
JAZ Proteins Interact with MYC5.
(A) Schematic diagrams show domain constructs of MYC5 and JAZ11. The diagrams display the conserved JAZ-interaction domain (JID; green), bHLH domain (orange), ZIM domain (purple), and Jas domain (blue). The numbers indicate positions of the first and last amino acids of the domain constructs.
(B) Y2H assay to test the interactions of 12 Arabidopsis JAZs with MYC5 and its related domains (as shown in [A]). JAZs were fused with the LexA DNA binding domain in pLexA, and MYC5 and its domains were fused with the activation domain (AD) in pB42AD. Interactions (represented by blue color) were assessed on 2% Gal/1% raffinose/SD/-Ura/-His/-Trp/-Leu/X-β-Gal medium.
(C) BiFC assay to detect the interactions of JAZ1 and JAZ10 with MYC5. JAZ1, JAZ10, and MYC5 were fused with the N-terminal fragment of YFP (nYFP) or the C-terminal fragment of YFP (cYFP) to generate JAZ1-nYFP, JAZ10-nYFP, and cYFP-MYC5. YFP fluorescence was detected 50 h after coexpression of the indicated construct pairs in leaves of N. benthamiana. The nuclei are indicated by 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) staining.
(D) Y2H assay to test the interactions of JAZ11 domain constructs (as shown in [A]) with MYC5. MYC5 was fused with the activation domain, and different JAZ11 domains were fused with the DNA binding domain.
The Subgroup IIIe bHLH Factors MYC2, MYC3, MYC4, and MYC5 Function Redundantly to Regulate Stamen Development and Seed Production
Subcellular localization analysis showed that MYC5 was localized in the nucleus (Supplemental Figure 2A), where JAZ proteins are also found (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). Expression pattern analysis with PMYC5-GUS (for β‑glucuronidase) reporter and quantitative real-time PCR showed that MYC5 was expressed in various tissues (Supplemental Figures 2B and 2C), with strong expression in flower and stamen (Supplemental Figure 2B), implying a possible role for MYC5 in the regulation of flower development. However, myc5 exhibited normal development in flower and stamen (Figure 2; Supplemental Figure 3).
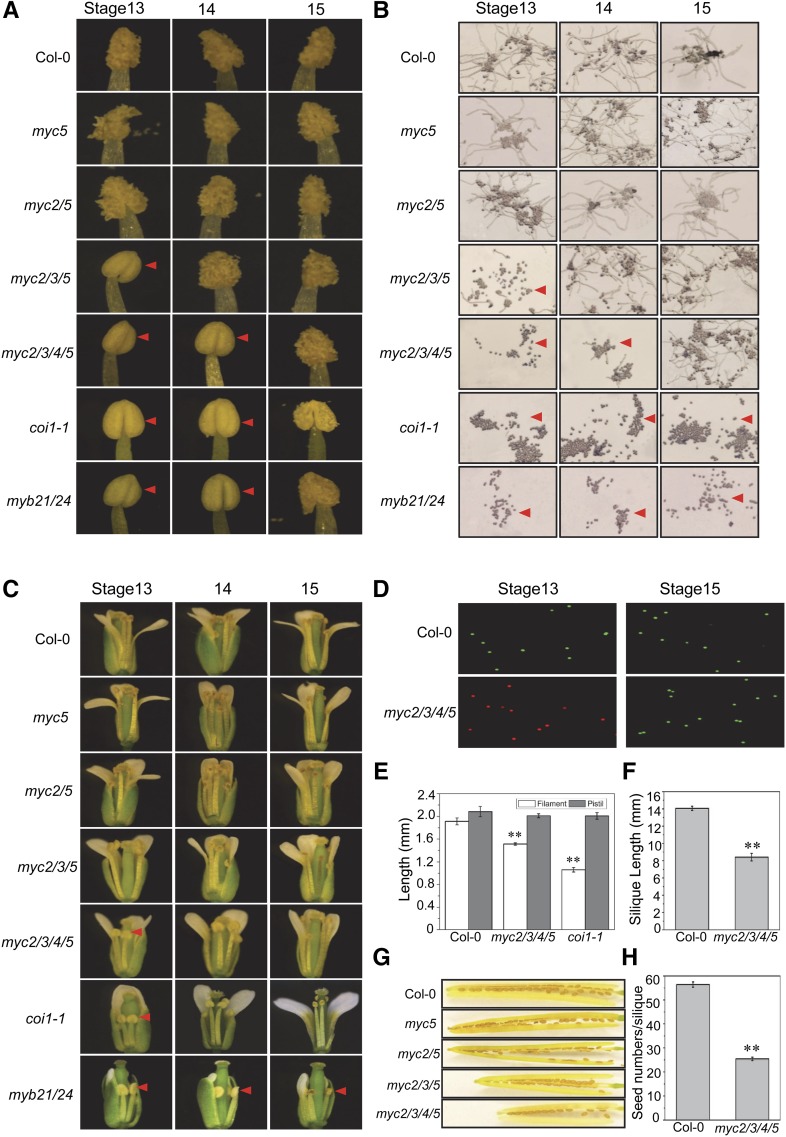
Figure 2.
The bHLH Subgroup IIIe Factors Function Redundantly to Regulate Stamen Development.
(A) Phenotypic observation of anther dehiscence of the indicated genotypes at floral stage 13, 14, or 15. Red arrowheads represent indehiscent anthers.
(B) In vitro germination of pollen grains from the flowers at floral stage 13, 14, or 15 in the indicated genotypes. Red arrowheads indicate pollen grains deficient in germination in vitro.
(C) Comparison of flowers at floral stage 13, 14, or 15 in the Col-0 wild type, myc5, myc2 myc5 (myc2/5), myc2 myc3 myc5 (myc2/3/5), myc2 myc3 myc4 myc5 (myc2/3/4/5), coi1-1, and myb21-3 myb24 (myb21/24). Red arrowheads indicate shorter filaments.
(D) Staining of pollen grains from Col-0 and myc2 myc3 myc4 myc5 (myc2/3/4/5) flowers at floral stage 13 or 15 with fluorescein diacetate and propidium iodide. Viable pollen grains are green, and unviable pollen grains are red.
(E) Filament length and pistil length at floral stage 13 in the indicated genotypes. Values are means ± se from three biological replicates. Asterisks represent Student’s t test significance compared with the Col-0 wild type (**P < 0.01).
(F) Silique length of Col-0 and myc2 myc3 myc4 myc5 (myc2/3/4/5). Values are means ± se from three biological replicates. Asterisks represent Student’s t test significance compared with the Col-0 wild type (**P < 0.01).
(G) Siliques from Col-0, myc5, myc2 myc5 (myc2/5), myc2 myc3 myc5 (myc2/3/5), and myc2 myc3 myc4 myc5 (myc2/3/4/5).
(H) Seed numbers per silique of Col-0 and myc2 myc3 myc4 myc5 (myc2/3/4/5). Values are means ± se from three biological replicates. Asterisks represent Student’s t test significance compared with the Col-0 wild type (**P < 0.01).
Previous phylogenetic analysis showed that the IIIe bHLH factors MYC5, MYC2, MYC3, and MYC4 share high identity and similarity at the amino acid level (Heim et al., 2003). We generated a series of the double, triple, and quadruple mutants through genetic crosses among myc2, myc3, myc4, and myc5 to examine whether MYC5 would exhibit redundancy with MYC2, MYC3, and MYC4.
Interestingly, compared with the wild type, all of the triple mutants, including myc2 myc3 myc4, myc2 myc3 myc5, myc2 myc4 myc5, and myc3 myc4 myc5, exhibited defects in stamen development at floral stage 13: their anthers failed to dehisce (Figure 2A; Supplemental Figure 4A), and the pollen grains were unviable and unable to germinate in vitro (Figure 2B; Supplemental Figure 4B). The anther dehiscence and pollen maturation were delayed in the triple mutants (Figure 2; Supplemental Figure 4). Furthermore, phenotypic comparison among these triple mutants showed that they exhibit similar severity in delayed stamen development, suggesting that MYC2, MYC3, MYC4, and MYC5 play equal roles in regulating stamen development. In contrast with the triple mutants, the single mutants and all the double mutants (myc2 myc5, myc3 myc5, myc4 myc5, myc2 myc3, myc2 myc4, and myc3 myc4) exhibited no obvious defects in stamen development (Figure 2; Supplemental Figures 4 and 5A).
Compared with the triple mutants, the myc2 myc3 myc4 myc5 quadruple mutant exhibited more severe defects in stamen development. In addition to the indehiscent anther and unviable pollen grains (Figures 2A, 2B, and 2D), the filament in myc2 myc3 myc4 myc5 did not elongate normally at floral stage 13 (Figures 2C and 2E), which is similar to coi1-1 and the myb21-3 myb24 double mutant (Figures 2A to 2C) (Song et al., 2011). At floral stage 14, in contrast with the normal stamen development in the wild type and the triple mutants, the anthers in the myc2 myc3 myc4 myc5 quadruple mutant failed to dehisce (similar to coi1-1 and myb21 myb24) (Figure 2A). The majority of the pollen grains in the quadruple mutant did not germinate in vitro (a few germinated pollen grains exhibited shorter pollen tubes) (Figure 2B). At floral stage 15, the anthers in myc2 myc3 myc4 myc5 dehisced (Figure 2A) and released viable pollen (Figures 2B and 2D), which ensured that myc2 myc3 myc4 myc5 was fertile and set seeds (Figures 2F to 2H).
Compared with the wild type, the siliques in the triple mutants contained somewhat fewer seeds (Figure 2G; Supplemental Figure 5B), while seed production was severely reduced in the myc2 myc3 myc4 myc5 quadruple mutant (Figures 2G and 2H).
Taken together (Figure 2; Supplemental Figures 4 and 5), these results demonstrated that the IIIe bHLH transcription factors MYC2, MYC3, MYC4, and MYC5 function redundantly to regulate stamen development and seed production.
Overexpression of MYC5 or MYC3 Can Partially Rescue Stamen Development in coi1-1
As the IIIe bHLH transcription factors act as targets of JAZ proteins (Figure 1) (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011) to mediate JA-regulated stamen development (Figure 2; Supplemental Figures 4 and 5), we next investigated whether the overexpression of MYC5 or MYC3 is able to overcome JAZ inhibition and rescue stamen development in the coi1-1 mutant, in which the JAZ repressors are highly accumulated (Chini et al., 2007; Thines et al., 2007).
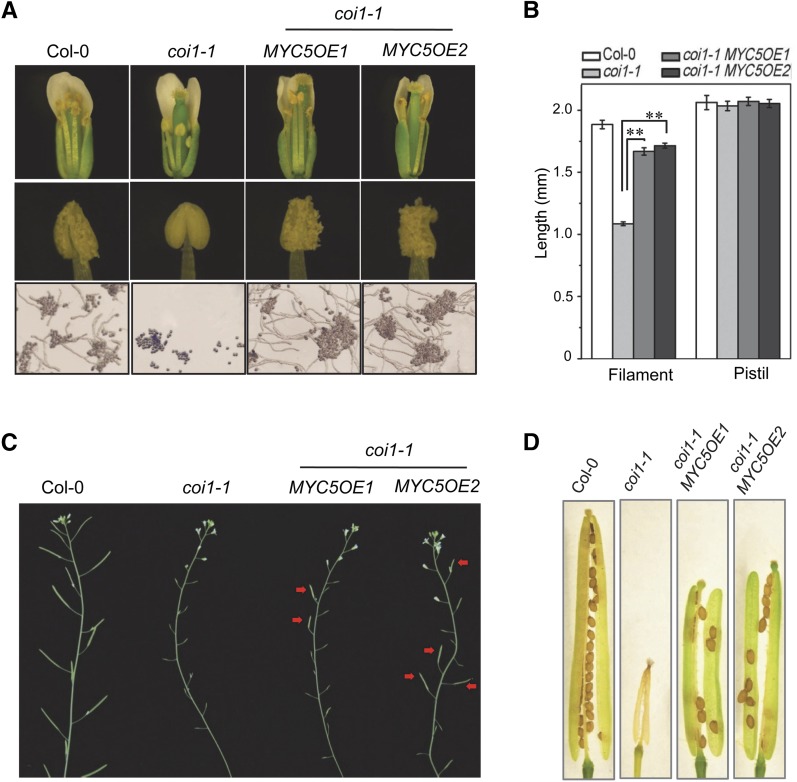
We generated coi1-1 plants overexpressing MYC5 (coi1-1 MYC5OE) and found that, among 42 individual transgenic lines, 4 lines showed partially recovered male fertility (2 representative lines are shown in Figure 3). In contrast with coi1-1, the transgenic plants coi1-1 MYC5OE1 and coi1-1 MYC5OE2 at floral stage 13 harbored fertile stamens, in which the filament could elongate and the anthers could dehisce to release viable pollen that could geminate in vitro (Figures 3A and 3B). The mature coi1-1 MYC5OE1 and coi1-1 MYC5OE2 plants could set small amounts of seed (Figures 3C and 3D).
Figure 3.
Overexpression of MYC5 Partially Restores Stamen Development in coi1-1.
(A) Comparison of flowers, anthers, and in vitro germination of pollen grains from flowers at floral stage 13 in the Col-0 wild type, coi1-1, and coi1-1 plants overexpressing MYC5 (coi1-1 MYC5OE1 and coi1-1 MYC5OE2).
(B) Filament length and pistil length in flowers at floral stage 13 from Col-0, coi1-1, and coi1-1 plants overexpressing MYC5 (coi1-1 MYC5OE1 and coi1-1 MYC5OE2). Values are means ± se from three biological replicates. Asterisks represent Student’s t test significance compared with coi1-1 (**P < 0.01).
(C) Main inflorescences in the indicated genotypes. Red arrows indicate fertile siliques in coi1-1 MYC5OE1 and coi1-1 MYC5OE2.
(D) Comparison of seed set in the indicated genotypes.
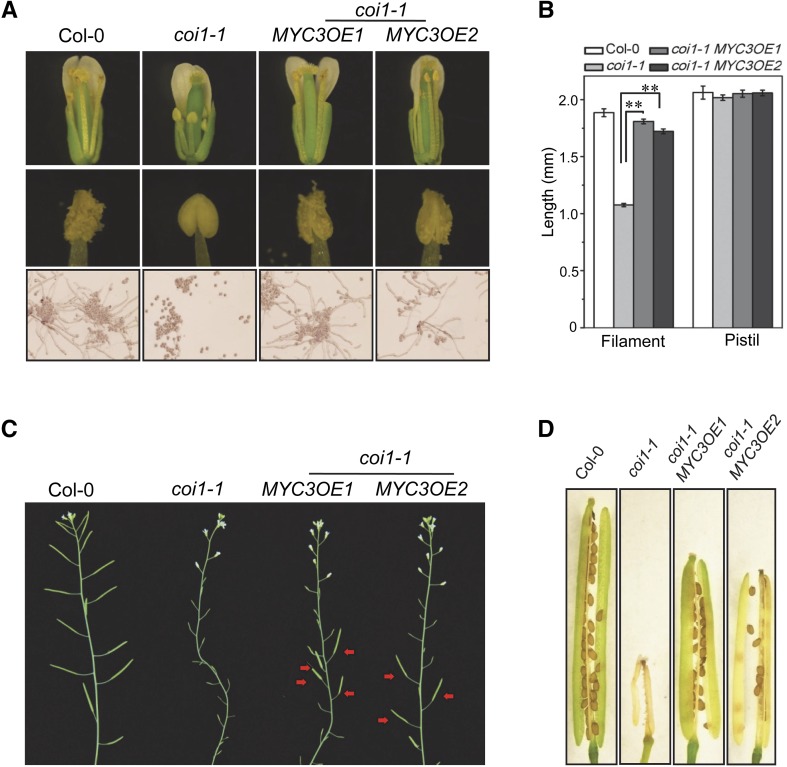
In the coi1-1 plants overexpressing MYC3 (coi1-1 MYC3OE), 4 out of 34 lines exhibited partially recovered stamen development (2 representative lines are shown in Figure 4). Taken together (Figures 3 and 4), these results demonstrate that high expression of MYC5 or MYC3 is able to relieve JAZ inhibition and partially recover stamen development in the coi1-1 mutant.
Figure 4.
Overexpression of MYC3 Partially Restores Stamen Development in coi1-1.
(A) Comparison of flowers, anthers, and in vitro germination of pollen grains from flowers at floral stage 13 in the Col-0 wild type, coi1-1, and coi1-1 plants overexpressing MYC3 (coi1-1 MYC3OE1 and coi1-1 MYC3OE2).
(B) Filament length and pistil length in flowers at floral stage 13 from Col-0, coi1-1, and coi1-1 plants overexpressing MYC3 (coi1-1 MYC3OE1 and coi1-1 MYC3OE2). Values are means ± se from three biological replicates. Asterisks represent Student’s t test significance compared with coi1-1 (**P < 0.01).
(C) Main inflorescences in the indicated genotypes. Red arrows indicate fertile siliques in coi1-1 MYC3OE1 and coi1-1 MYC3OE2.
(D) Comparison of seed set in the indicated genotypes.
MYC2, MYC3, MYC4, and MYC5 Are Required for JA-Inducible Expression of MYB21, MYB24, MYB57, and MYB108
Previous studies showed that JA induces the expression of the R2R3 MYB transcription factors MYB21, MYB24, MYB57, and MYB108, which are essential for JA-regulated stamen development (Mandaokar et al., 2006; Cheng et al., 2009; Mandaokar and Browse, 2009; Song et al., 2011). We further examined whether the IIIe bHLH factors MYC2, MYC3, MYC4, and MYC5 are required for the JA-inducible expression of these R2R3 MYB transcription factors.
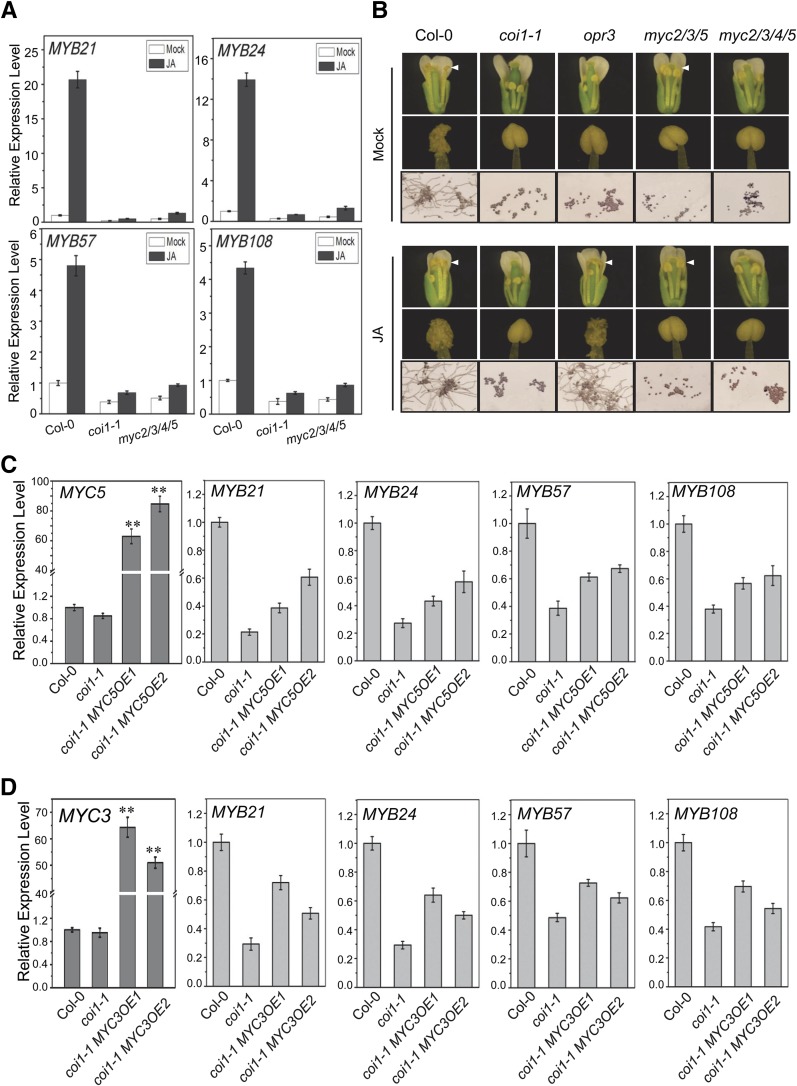
As shown in Figure 5A, the expression of MYB21, MYB24, MYB57, and MYB108 was partially decreased in the flower buds of coi1-1 and the myc2 myc3 myc4 myc5 quadruple mutant compared with that of the wild type, suggesting that COI1 and the IIIe bHLH factors are required for the expression of MYB21, MYB24, MYB57, and MYB108.
Figure 5.
MYC2, MYC3, MYC4, and MYC5 Are Required for JA-Inducible Expression of MYB21, MYB24, MYB57, and MYB108.
(A) RT-qPCR analysis for MYB21, MYB24, MYB57, and MYB108 in young flower buds of the Col-0 wild type, coi1-1, and myc2 myc3 myc4 myc5 (myc2/3/4/5) treated without (Mock) or with methyl jasmonate (JA). ACTIN8 was used as the internal control. Values are means ± se from three biological replicates.
(B) Comparison of flowers, anther dehiscence, and in vitro germination of pollen grains at floral stage 13 in Col-0, coi1-1, opr3, myc2 myc3 myc5 (myc2/3/5), and myc2 myc3 myc4 myc5 (myc2/3/4/5) treated without (Mock) or with methyl jasmonate (JA). White arrowheads indicate fully elongated filaments.
(C) and (D) RT-qPCR analysis of gene expression in Col-0, coi1-1, and coi1-1 plants overexpressing MYC5 (coi1-1 MYC5OE1 and coi1-1 MYC5OE2) (C) or coi1-1 plants overexpressing MYC3 (coi1-1 MYC3OE1 and coi1-1 MYC3OE2) (D). Expression of MYC5 (C) and MYC3 (D) was detected in 3-week-old plants, and expression of MYB21, MYB24, MYB57, and MYB108 was tested in young flower buds of the indicated genotypes using ACTIN8 as the internal control. Values are means ± se from three biological replicates. Asterisks represent Student’s t test significance compared with the Col-0 wild type (**P < 0.01).
Moreover, JA treatment can induce the expression of MYB21, MYB24, MYB57, and MYB108 in the flower buds of the wild type (Figure 5A). In contrast with the JA-treated wild type, we found that, similar to coi1-1, the myc2 myc3 myc4 myc5 quadruple mutant was clearly insensitive to the JA-inducible expression of the R2R3 MYB transcription factors (Figure 5A). Consistent with these results, JA treatment could not rescue stamen development in myc2 myc3 myc4 myc5 and coi1-1 (Figure 5B), as indicated by filament elongation, anther dehiscence, and pollen germination in vitro at floral stage 13, while the application of JA was able to rescue stamen development in the JA-deficient mutant opr3 (Figure 5B).
Furthermore, we found that the overexpression of MYC5 or MYC3, which partially restored stamen development in coi1-1 (Figures 3 and 4), mildly but clearly restored the expression of MYB21, MYB24, MYB57, and MYB108 in fertile young flower buds of the coi1-1 MYC5OE and coi1-1 MYC3OE plants (Figures 5C and 5D). In addition, we applied the ERF-associated amphiphilic repression (EAR) motif-based chimeric repressor-silencing technology (Hiratsu et al., 2003) to overexpress the MYC5 chimeric repressors (MYC5-EAR) in wild-type plants, which disrupted stamen development, abolished male fertility, and suppressed the expression of MYB21, MYB24, MYB57, and MYB108 in the flower buds of the sterile MYC5-EAR plants (Supplemental Figure 6). Our results are consistent with similar observations of MYC5-SRDX transgenic plants reported recently (Figueroa and Browse, 2015).
Taken together (Figures 2 to 5; Supplemental Figures 4 and 5), these results demonstrated that the IIIe bHLH factors MYC2, MYC3, MYC4, and MYC5 function redundantly to regulate stamen development and mediate the JA-inducible expression of the MYB genes (MYB21, MYB24, MYB57, and MYB108).
The IIIe bHLH Factors Interact with the MYB Transcription Factors MYB21 and MYB24
Having showed that the expression of these R2R3-MYB factors is regulated by the IIIe bHLH factors (MYC2, MYC3, MYC4, and MYC5) (Figure 5), we next investigated whether the R2R3-MYB factors, such as MYB21 and MYB24, interact with the IIIe bHLH factors.
We used MYC5 as the representative and examined its interaction with MYB21 and MYB24. As both MYB21 and MYB24 exhibited autoactivation in the Y2H system, we used MYB21NT and MYB24NT with deletion of the C-terminal activation domain (NYWG/SM/VDDI/LWS/P motif) to examine their interaction with MYC5 in the Y2H system (Figure 6A). As shown in Figure 6B, both MYB21NT and MYB24NT interacted with MYC5 in yeast. Moreover, MYB21NT and MYB24NT interacted with MYC5NT but not with MYC5CT (Figure 6B). We next performed coimmunoprecipitation (Co-IP) assays to test the interaction of MYC5 with MYB21 in planta. Our Co-IP assays showed that flag-MYC5 could coimmunoprecipitate with myc-MYB21 (Figure 6C).
Figure 6.
MYC2, MYC3, MYC4, and MYC5 Interact with MYB21 and MYB24.
(A) Schematic diagrams of MYB21, MYB24, and their N-terminal domains (MYB21NT and MYB24NT). MYB21NT and MYB24NT contain the conserved R2 (red) and R3 (blue) domains but not the NYWG/SM/VDDI/LWS/P motif (orange).
(B) Y2H assay to show interactions between MYB21NT/MYB24NT and MYC5. MYB21NT and MYB24NT were fused with the DNA binding domain (BD), and MYC5 was fused with the activation domain (AD).
(C) Co-IP assay to verify the interaction between MYC5 and MYB21 in vivo. Flag-MYC5 was coexpressed without (empty control) or with myc-MYB21 or myc-COI1 (negative control) in N. benthamiana leaves. The total protein extracts from N. benthamiana leaves transiently expressing flag-MYC5, flag-MYC5 plus myc-MYB21, or flag-MYC5 plus myc-COI1 were immunoprecipitated using the anti-c-myc antibody-conjugated agarose and were blotted with anti-flag or anti-c-myc antibody.
(D) BiFC assay to detect interactions of MYC2, MYC3, MYC4, and MYC5 with MYB21 and MYB24. MYC2, MYC3, MYC4, and MYC5 were fused with nYFP, and MYB21 and MYB24 were fused with cYFP. The nuclei are indicated by 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) staining.
We also employed BiFC assays to verify the interaction of MYC5 with MYB21 and MYB24 in planta. The coexpression of MYC5-nYFP with cYFP-MYB21 or cYFP-MYB24 in Nicotiana benthamiana leaves produced strong YFP signals (Figure 6D), but the negative controls did not (Supplemental Figure 1), suggesting that MYC5 interacts with MYB21 and MYB24 in planta. Similar results were observed for the coexpression of MYC2-nYFP, MYC3-nYFP, and MYC4-nYFP with cYFP-MYB21 or cYFP-MYB24 (Figure 6D; Supplemental Figure 1).
Taken together (Figure 6), these Y2H, Co-IP, and BiFC assays demonstrate that IIIe bHLH factors (MYC2, MYC3, MYC4, and MYC5) interact with MYB21 and MYB24 to form the bHLH-MYB transcription complex.
JAZ1 Attenuates the Transcriptional Activation Function of the bHLH-MYB Complex
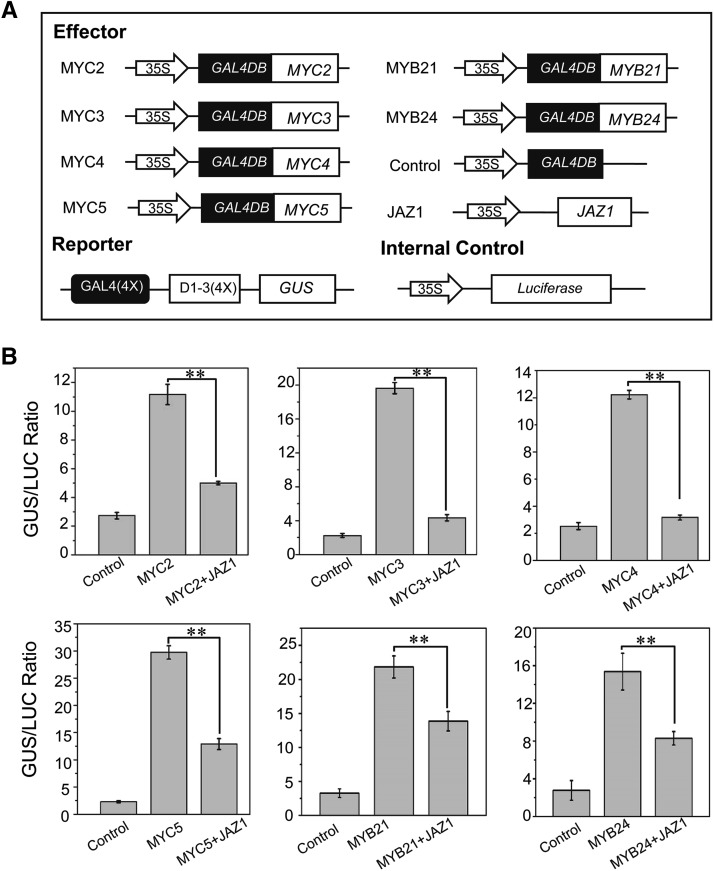
Since the IIIe bHLH proteins (MYC2, MYC3, MYC4, and MYC5) and the MYB proteins (MYB21 and MYB24) are targets of JAZ proteins, we examined whether the JAZ interactions repress the transcriptional activation activity of these components.
The fusion of MYC2 with GAL4DB (GAL4DB-MYC2) driven by the 35S promoter was used as an effector, and the GUS gene under the control of four copies of GAL4 DNA binding site [GAL4(4x)-D1-3(4x)] was used as a reporter, whereas the luciferase (LUC) gene driven by the 35S promoter was used as the internal control (Figure 7A). The expression of GAL4DB-MYC2 clearly increased the GUS:LUC ratio, while coexpression of JAZ1 with GAL4DB-MYC2 clearly decreased the GUS:LUC ratio (Figure 7B; Supplemental Figure 7). The results suggested that MYC2 acts as a transcriptional activator, while JAZ1 attenuates the transcriptional activation function of MYC2. Similarly, we observed that JAZ1 can inhibit the transcriptional activity of MYC3, MYC4, MYC5, MYB21, and MYB24 (Figure 7).
Figure 7.
JAZ1 Attenuates the Transcriptional Activation Function of MYC2, MYC3, MYC4, MYC5, MYB21, and MYB24.
(A) Schematic diagrams of the constructs used in the transient expression assays in (B).
(B) Transient expression assays show that JAZ1 inhibits the transcriptional activation function of MYC2, MYC3, MYC4, MYC5, MYB21, and MYB24. Values are means ± se from three biological replicates. Asterisks represent Student’s t test significance between pairs indicated with brackets (**P < 0.01).
In summary, these results (Figures 1 and 7) suggest that JAZ proteins repress the transcriptional activation function of the IIIe bHLH (MYC2, MYC3, MYC4, and MYC5) and MYB (MYB21 and MYB24) components of the bHLH-MYB complex.
MYC2, MYC3, MYC4, and MYC5 Function Cooperatively with MYB21 and MYB24 to Regulate Stamen Development and Seed Production
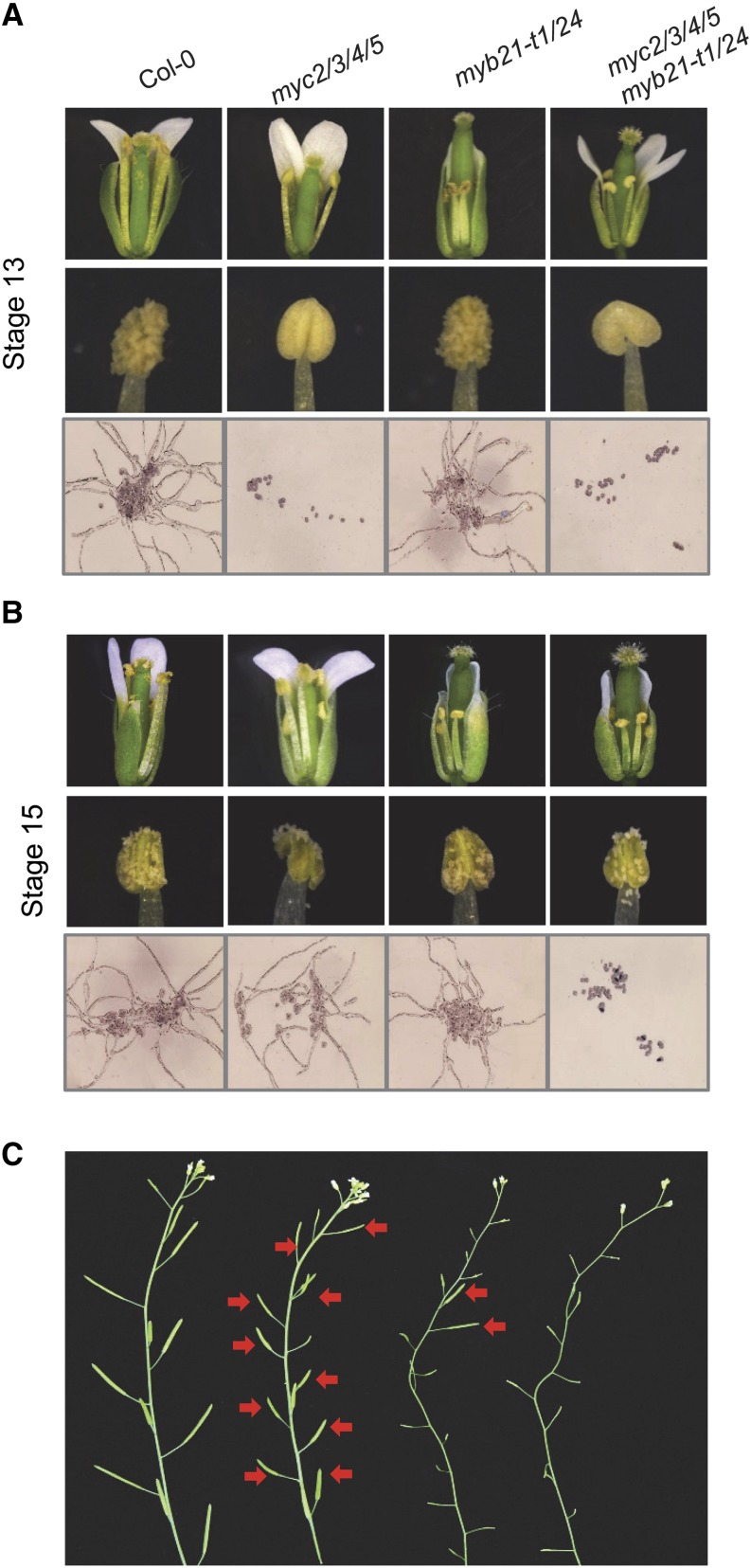
To genetically investigate whether the IIIe bHLH factors (MYC2, MYC3, MYC4, and MYC5) cooperate with MYB21 and MYB24 to regulate stamen development, we used the myc2 myc3 myc4 myc5 quadruple mutant and the myb21-t1 myb24 double mutant (with a weak allele for myb21) (Cheng et al., 2009) to generate the myc2 myc3 myc4 myc5 myb21-t1 myb24 sextuple mutant.
As shown in Figure 8, the myc2 myc3 myc4 myc5 myb21-t1 myb24 sextuple mutant plants were completely male-sterile: they exhibited short filaments, failure of anther dehiscence, and unviable pollen at the late floral stage (Figures 8A to 8C). By contrast, myc2 myc3 myc4 myc5 and myb21-t1 myb24 harbored fertile anthers at floral stage 15 and exhibited partial fertility (Figures 8A to 8C). These results demonstrated that the IIIe bHLH factors (MYC2, MYC3, MYC4, and MYC5) and the MYB factors (MYB21 and MYB24) cooperatively mediate JA-regulated stamen development and seed production.
Figure 8.
The bHLH and MYB Factors Cooperatively Mediate JA-Regulated Stamen Development.
(A) and (B) Phenotypes of flowers, stamens, and in vitro pollen germination in the Col-0 wild type, myc2 myc3 myc4 myc5 (myc2/3/4/5), myb21-t1 myb24, and myc2 myc3 myc4 myc5 myb21-t1 myb24 (myc2/3/4/5 myb21-t1/24) at floral stage 13 (A) and floral stage 15 (B).
(C) Inflorescences in the indicated genotypes. Red arrows indicate fertile siliques of myc2 myc3 myc4 myc5 and myb21-t1 myb24.
DISCUSSION
We previously revealed that JAZ repressors interact with and repress the MYB transcription factors MYB21, MYB24, and MYB57 to modulate JA-regulated stamen development (Song et al., 2011). In this study, we elucidated that the IIIe bHLH transcription factor MYC5 acts as a target of JAZ repressors to function redundantly with the other IIIe bHLH factors MYC2, MYC3, and MYC4 in the regulation of stamen development and seed production. Moreover, these IIIe bHLH factors interact with the MYB transcription factors MYB21 and MYB24 to form the bHLH-MYB transcription complex and cooperatively regulate stamen development.
Our results suggest that, as diagrammed in Figure 9, JAZ repressors interact with and repress the bHLH-MYB complex to suppress stamen development. In response to environmental cues or developmental signals (such as flowering), plants biosynthesize JA that is perceived by its receptor COI1 to induce JAZ degradation and release the bHLH-MYB complex, which subsequently regulates the expression of downstream genes (as well as some members of the bHLH-MYB complex) to mediate stamen development. Based on our current findings together with our previous observations (Cheng et al., 2009), we speculate that gibberellin also acts through the bHLH-MYB complex to regulate stamen development, although additional unidentified components are probably involved: DELLAs inhibit the expression of JA biosynthetic genes and repress the biosynthesis of JA in flowers, thereby leading to the suppression of stamen development. DELLAs directly interact with and repress MYC2 (Hong et al., 2012), and it will be interesting to investigate whether the DELLA proteins directly target all the bHLH members (MYC2, MYC3, MYC4, and MYC5) and MYB members (MYB21 and MYB24) to inhibit the transcriptional function of the bHLH-MYB complex for the regulation of gibberellin-regulated stamen development.
Figure 9.
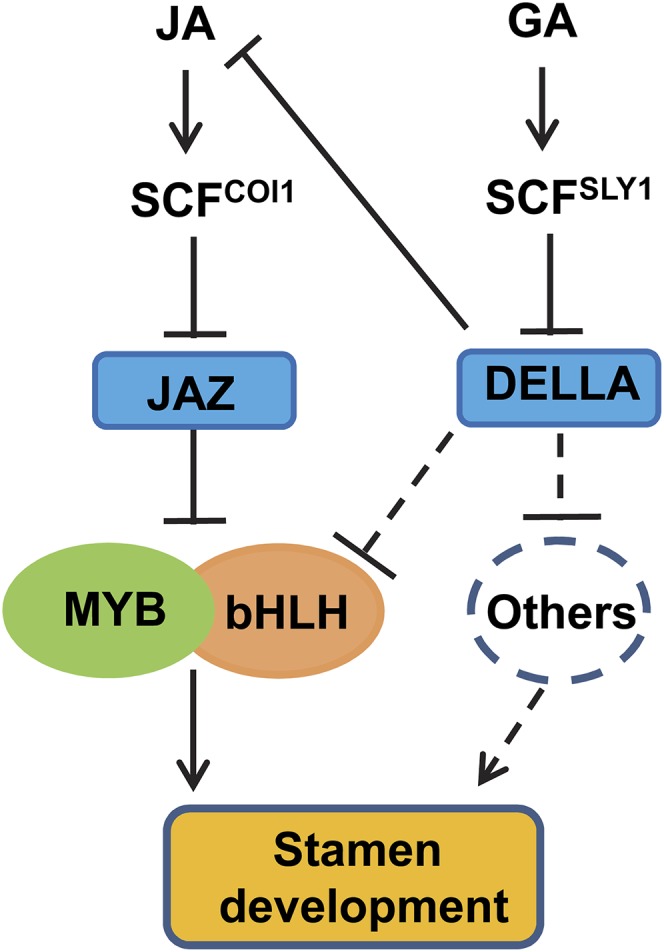
A Simplified Model for JA-Regulated Stamen Development.
JAZ repressors interact with and repress the bHLH-MYB complex consisting of the IIIe bHLH factors MYC2, MYC3, MYC4, and MYC5 (indicated as bHLH) and the R2R3 MYB factors MYB21 and MYB24 (indicated as MYB). JA is perceived by COI1 to induce JAZ degradation and release the bHLH-MYB complex, which regulates the expression of downstream genes (as well as some members of the bHLH-MYB complex) to mediate stamen development. Gibberellin (GA) may also act through the bHLH-MYB complex to regulate stamen development: DELLAs repress the biosynthesis of JA in flowers (Cheng et al. 2009) to suppress JA-regulated stamen development. In addition, DELLAs directly interact with and repress MYC2 (Hong et al. 2012). Further experiments are required to verify whether the interaction of DELLAs with MYC2 or other unidentified components (indicated as Others) modulates gibberellin-regulated stamen development.
We observed that the MYC members positively regulate the expression of the MYB genes: the expression of MYB21, MYB24, and MYB57 was downregulated in the flower buds of the myc2 myc3 myc4 myc5 quadruple mutant (Figure 5A), and conversely, overexpression of MYC3 or MYC5 partially restored the expression of MYB21, MYB24, and MYB57 in the flower buds of coi1-1 (Figures 5C and 5D). Interestingly, the expression of MYC2 was obviously increased in the flower buds of the myb21 myb24 double mutants, while the expression of MYC5 was not elevated (Supplemental Figure 8A). A previous study showed that myb21 myb24 exhibits elevated JA biosynthesis in flowers (Reeves et al., 2012). It is likely that the elevated JA biosynthesis in myb21 myb24 may upregulate the expression of MYC2 but not MYC5. Consistent with this idea, we found that treatment with JA significantly induced the expression of MYC2 but not MYC5 (Supplemental Figures 8B and 8D). The feedback regulation among the MYCs and MYBs is complicated and requires further experimental verification.
Previous research has shown that bHLH factors (GL3, EGL3, or TT8) interact with R2R3 MYB factors (MYB75 or GL1), together with the WD-repeat protein TTG1, to form a transcription complex, referred to as the WD-repeat/bHLH/MYB complex (Zhang et al., 2003). Within this complex, the bHLH factor (such as GL3) interacts with the MYB factors (MYB75 or GL1) via their N-terminal domains, while JAZ repressors competitively interact with the N-terminal domains of these bHLH and MYB factors to disrupt and repress the WD-repeat/bHLH/MYB complex essential for JA-regulated anthocyanin biosynthesis and trichome initiation (Qi et al., 2011, 2014). JA induces the degradation of JAZs to release this WD-repeat/bHLH/MYB complex, which further activates the expression of the downstream genes to enhance anthocyanin biosynthesis and trigger trichome initiation (Qi et al., 2011).
Analogously to the WD-repeat/bHLH/MYB complex, in our newly identified bHLH-MYB transcription complex, a similar N-terminal interaction also mediates the association of the bHLH factors (MYC2, MYC3, MYC4, and MYC5) with the MYB factors (MYB21 and MYB24). JAZ proteins similarly interact with (Figure 1) (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011; Song et al., 2011) and repress these bHLH factors and MYB factors (Figure 7) to disrupt the bHLH-MYB complex for the modulation of JA-regulated stamen development. Although interactions occur among multiple bHLH factors (or MYB members) (Fernández-Calvo et al., 2011; Song et al., 2011), it is still unclear whether the bHLH-MYB complex contains a single or multiple bHLH factors (or MYB factors) in planta.
TTG1, a member of the WD-repeat family, acts as a key component of the WD-repeat/bHLH/MYB complex to regulate anthocyanin biosynthesis and trichome initiation (Zhang et al., 2003). Several target genes are directly activated by the WD-repeat/bHLH/MYB complex for the regulation of anthocyanin biosynthesis and trichome initiation (Zhang et al., 2003; Gonzalez et al., 2008; Morohashi and Grotewold, 2009). It remains to be investigated whether, in our newly identified complex, another unidentified WD-repeat protein similarly associates with the IIIe bHLH factors (MYC2, MYC3, MYC4, and MYC5) and MYB factors (MYB21 and MYB24) to regulate stamen development.
The bHLH factors MYC2, MYC3, and MYC4 are known to interact with each other to form homodimers/heterodimers and to associate with MYB factors (MYB28, MYB29, MYB34, MYB51, MYB76, and MYB122) to regulate glucosinolate biosynthesis (Fernández-Calvo et al., 2011; Schweizer et al., 2013). This study demonstrated that MYC factors (MYC2, MYC3, MYC4, and MYC5) interact with MYB members (MYB21 and MYB24) to form a bHLH-MYB complex for the regulation of stamen development. Identification and characterization of target genes for the bHLH-MYB complex will provide insights into the molecular mechanism underlying JA-regulated stamen development.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana mutants coi1-1 (Xie et al., 1998), opr3 (Stintzi and Browse, 2000), and myc2-2 (Salk_083483) (Boter et al., 2004) were described previously. myc3 (GK445B11), myc4 (GK491E10), and myc5 (Salk_060048C) were obtained from the NASC or ABRC. The double, triple, and quadruple mutants of the bHLH subgroup IIIe factors were generated by genetic crossing with myc2-2, myc3, myc4, and myc5.
Arabidopsis thaliana seeds were sterilized with 20% bleach, plated on Murashige and Skoog medium (Sigma-Aldrich), chilled at 4°C for 3 d, and transferred to a growth room under a 16-h (20 to 24°C)/8-h (16 to 19°C) light/dark condition as described previously (Song et al., 2014a). Nicotiana benthamiana was grown in a growth room under a 16-h (25 to 28°C)/8-h (22 to 25°C) light/dark condition as described previously (Song et al., 2014a).
Y2H Screening and Y2H Assays
The Y2H screening method was described previously (Song et al., 2013a). For the Y2H assay, JAZ1, JAZ2, JAZ3, JAZ4, JAZ5, JAZ6, JAZ7, JAZ8 JAZ9, JAZ10, JAZ11, JAZ12, and MYC5, or the related domains of JAZ11, MYC5, MYB21, and MYB24, were inserted into pLexA or pB42AD vector, respectively. Primers used for the vector construction are listed in Supplemental Table 1. The methods for yeast transformation and the interaction assay using EGY48 were as described previously (Song et al., 2011). Y2H images were taken 3 d after incubation.
Co-IP Assay
N. benthamiana leaves were infiltrated with Agrobacterium tumefaciens strains harboring flag-MYC5, flag-MYC5 plus myc-MYB21, or flag-MYC5 plus myc-COI1 (Xu et al., 2002). At 50 h after infiltration, 3 g of Agrobacterium-infiltrated leaves for each combination was collected. The total protein was extracted with Co-IP buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM DTT, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride, 50 mM MG132, and complete protease inhibitor cocktail [Roche]) and centrifuged twice at 16,000g at 4°C. The supernatant was concentrated to 400 mL and incubated with the agarose-conjugated anti-myc matrix (Abmart) for 2 h with rotation at 4°C. After washing three times with 1 mL of immunoprecipitation buffer, the agarose beads were denatured in 100 mL of SDS loading buffer. The samples were separated by SDS-PAGE and immunoblotted with the corresponding antibody.
BiFC Assays
For BiFC assays, the coding sequences of Arabidopsis JAZ1, JAZ10, MYC2, MYC3, MYC4, MYC5, MYB21, and MYB24 were inserted individually into the nYFP or cYFP vector through Gateway methods (Invitrogen) (Qi et al., 2011; Song et al., 2011). Primer pairs for vector construction are listed in Supplemental Table 1. The Agrobacterium strains containing the indicated vectors were coinfiltrated into leaves of N. benthamiana as described previously (Song et al., 2011; Qi et al., 2013). After infiltration, plants were placed at 24°C for 50 h before observation.
Reverse Transcription and Quantitative Real-Time PCR
For Supplemental Figure 2C, the expression of MYC5 was analyzed in root, stem, rosette leaf, stem leaf, and flower from 5-week-old Arabidopsis plants. For Figures 5C and 5D and Supplemental Figure 6D, the expression of MYC3 and MYC5 was analyzed in 3-week-old Arabidopsis plants. The expression of MYB21, MYB24, MYB57, and MYB108 in Figures 5C and 5D and Supplemental Figure 6E and the expression of MYC2, MYC3, and MYC5 in Supplemental Figure 8A were analyzed in young flower buds from 5-week-old Arabidopsis plants. For Figure 5A and Supplemental Figures 8B to 8D, the expression of MYB21, MYB24, MYB57, MYB108, MYC2, MYC3, and MYC5 was analyzed in young flower buds from 5-week-old Arabidopsis plants mock treated or treated with 100 μM methyl jasmonate for 4 h. The materials were harvested for RNA extraction, subsequent reverse transcription, and quantitative real-time PCR analysis for the indicated genes. Quantitative real-time PCR analysis was performed with the RealMasterMix (SYBR Green I; Takara) using the ABI7500 real-time PCR system as described previously (Song et al., 2013a). ACTIN8 was used as the internal control. The primers for reverse transcription and quantitative real-time PCR (RT-qPCR) analysis are listed in Supplemental Table 2.
Transcriptional Activity Assays
For Arabidopsis protoplast transient expression assays, MYC2, MYC3, MYC4, MYC5, MYB21, and MYB24 were fused with the GAL4DB under the control of the 35S promoter, and JAZ1 was inserted into the pGreenII 62-SK vector under the control of the 35S promoter (Hellens et al., 2005). The GUS gene driven by four copies of upstream GAL4 DNA binding sites [GAL4(4x)-D1-3(4x)] was used as a reporter construct (Tiwari et al., 2001), and LUC under the control of the 35S promoter was used as the internal control. Preparation of Arabidopsis mesophyll protoplasts and transfection were performed as described previously (Yoo et al., 2007). GUS:LUC ratios are presented. Primers used for plasmid construction are shown in Supplemental Table 1.
Generation of Transgenic Plants
To generate Arabidopsis MYC3 or MYC5 overexpression constructs, the coding sequence of MYC3 or MYC5 was inserted into modified pCAMBIA1300 vector under the control of the 35S promoter. These constructs were transformed into Arabidopsis COI1/coi1-1 plants using the Agrobacterium-mediated floral dip method. To generate Arabidopsis EAR domain-fused MYC5 (MYC5-EAR) overexpression constructs, the coding sequence of MYC5 was inserted into modified pCAMBIA1300 with fusion of the EAR motif (GLDLDLELRLGFA) under the control of the 35S promoter. The construct was transformed into Arabidopsis Columbia-0 (Col-0) wild-type plants using the Agrobacterium-mediated floral dip method. Primer pairs for vector construction are listed in Supplemental Table 1.
GUS Staining
An ∼1795-bp promoter region of MYC5 was amplified and inserted into pCAMBIA1391Z vector to drive the GUS reporter gene (PMYC5-GUS). The construct was transformed into Arabidopsis plants using the Agrobacterium-mediated floral dip method. Roots, seedlings, leaves, inflorescences, flowers, and stamens from the plants transgenic for PMYC5-GUS were used for histochemical staining of GUS as described previously (Song et al., 2013a).
Measurement of Pistil and Filament Length
To measure the filament and pistil length, 10 flowers at floral stage 13 (Smyth et al., 1990) for each genotype were harvested, and the pistil and one of the four longer filaments for each flower were measured using a microscope.
Pollen Germination Assays
Pollen germination assays were performed as described previously (Boavida and McCormick, 2007). Pollen grains at the indicated floral stage (Smyth et al., 1990) were dipped into the pollen germination medium (10% [w/v] Suc, 0.01% boric acid, 1 mM MgSO4, 5 mM CaCl2, 5 mM KCl, pH 7.5, and 1.5% agar), incubated at 22°C in the dark for 10 h, and finally observed using a microscope.
Subcellular Localization
The coding sequence of MYC5 was inserted into pEGAD vector for fusion with GFP under the control of the 35S promoter to generate the GFP-MYC5 construct. Agrobacterium containing the GFP-MYC5 or GFP construct was resuspended in infiltration buffer and infiltrated into N. benthamiana leaves by a needleless syringe. After infiltration, plants were placed at 24°C for 50 h before GFP observation.
Accession Numbers
The Arabidopsis Genome Initiative numbers for the genes mentioned in this article are as follows: COI1 (AT2G39940), JAZ1 (AT1G19180), JAZ2 (AT1G74950), JAZ3 (AT3G17860), JAZ4 (AT1G48500), JAZ5 (AT1G17380), JAZ6 (AT1G72450), JAZ7 (AT2G34600), JAZ8 (AT1G30135), JAZ9 (AT1G70700), JAZ10 (AT5G13220), JAZ11 (AT3G43440), JAZ12 (AT5G20900), MYC2 (AT1G32640), MYC3 (AT5G46760), MYC4 (AT4G17880), MYC5 (AT5G46830), MYB21 (AT3G27810), MYB24 (AT5G40350), MYB57 (AT3G01530), MYB108 (AT3G06490), and ACTIN8 (AT1G49240).
Supplemental Data
Supplemental Figure 1. Negative Controls for the BiFC Experiments.
Supplemental Figure 2. Subcellular Localization and Expression Pattern of MYC5.
Supplemental Figure 3. RT-qPCR Analysis of MYC5 Expression in the T-DNA Insertion Mutant.
Supplemental Figure 4. The bHLH Subgroup IIIe Factors Function Redundantly to Regulate Anther Dehiscence and Germinating Ability of Pollen.
Supplemental Figure 5. The bHLH Subgroup IIIe Factors Function Redundantly to Regulate Filament Elongation and Seed Set.
Supplemental Figure 6. Overexpression of MYC5-EAR Leads to Male Sterility.
Supplemental Figure 7. JAZ1 Cannot Attenuate the Basal Activity of GAL4DB.
Supplemental Figure 8. Expression Level of MYC2, MYC3, and MYC5 in myb21-3 myb24 and coi1-1 Mutants.
Supplemental Table 1. Primers Used for Vector Construction.
Supplemental Table 2. Primers Used for RT-qPCR Analysis.
Supplementary Material
Acknowledgments
We thank John Browse, Roberto Solano, Chuanyou Li, Jinrong Peng, and Kang Chong for materials. This work was supported by the Ministry of Science and Technology of China (973 Program Grant 2011CB915404) and the National Natural Science Foundation of China (Grants 31230008, 91417302, 31421001, and 31400253).
AUTHOR CONTRIBUTIONS
T.Q., H.H., S.S., and D.X. designed the research. T.Q., H.H., and S.S. performed research. T.Q., H.H., S.S., and D.X. analyzed data. T.Q., S.S., and D.X. wrote the article.
Glossary
- bHLH
basic helix-loop-helix
- JA
jasmonate
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- Co-IP
coimmunoprecipitation
- RT-qPCR
reverse transcription and quantitative real-time PCR
- Col-0
Columbia-0
References
- Acosta I.F., Laparra H., Romero S.P., Schmelz E., Hamberg M., Mottinger J.P., Moreno M.A., Dellaporta S.L. (2009). tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323: 262–265. [DOI] [PubMed] [Google Scholar]
- Boavida L.C., McCormick S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52: 570–582. [DOI] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205. [DOI] [PubMed] [Google Scholar]
- Cai Q., Yuan Z., Chen M., Yin C., Luo Z., Zhao X., Liang W., Hu J., Zhang D. (2014). Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5: 3476. [DOI] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Sun L., Qi T., Zhang B., Peng W., Liu Y., Xie D. (2011). The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4: 279–288. [DOI] [PubMed] [Google Scholar]
- Chico J.M., Fernández-Barbero G., Chini A., Fernández-Calvo P., Díez-Díaz M., Solano R. (2014). Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26: 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Chung H.S., Cooke T.F., Depew C.L., Patel L.C., Ogawa N., Kobayashi Y., Howe G.A. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geyter N., Gholami A., Goormachtig S., Goossens A. (2012). Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 17: 349–359. [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer E.E., Mueller M.J. (2013). ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 64: 429–450. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male-sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P., Browse J. (2015). Male sterility in Arabidopsis induced by overexpression of a MYC5-SRDX chimeric repressor. Plant J. 81: 849–860. [DOI] [PubMed] [Google Scholar]
- Fonseca S., Fernández-Calvo P., Fernández G.M., Díez-Díaz M., Gimenez-Ibanez S., López-Vidriero I., Godoy M., Fernández-Barbero G., Van Leene J., De Jaeger G., Franco-Zorrilla J.M., Solano R. (2014). bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS ONE 9: e86182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53: 814–827. [DOI] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Hong G.J., Xue X.Y., Mao Y.B., Wang L.J., Chen X.Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66. [DOI] [PubMed] [Google Scholar]
- Huang H., Wang C., Tian H., Sun Y., Xie D., Song S. (2014). Amino acid substitutions of GLY98, LEU245 and GLU543 in COI1 distinctively affect jasmonate-regulated male fertility in Arabidopsis. Sci. China Life. Sci. 57: 145–154. [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Lyons R. (2014). Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26: 2285–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17: 22–31. [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56: 393–434. [DOI] [PubMed] [Google Scholar]
- Mandaokar A., Browse J. (2009). MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 149: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A., Thines B., Shin B., Lange B.M., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46: 984–1008. [DOI] [PubMed] [Google Scholar]
- McConn M., Browse J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Grotewold E. (2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Halitschke R., Kim H.B., Baldwin I.T., Feldmann K.A., Feyereisen R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31: 1–12. [DOI] [PubMed] [Google Scholar]
- Pauwels L., Goossens A. (2011). The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett A.R., Thomas S.G., Wilson Z.A., Hedden P. (2011). Gibberellin control of stamen development: A fertile field. Trends Plant Sci. 16: 568–578. [DOI] [PubMed] [Google Scholar]
- Qi T., Huang H., Wu D., Yan J., Qi Y., Song S., Xie D. (2014). Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26: 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Song S., Xie D. (2013). Modified bimolecular fluorescence complementation assay to study the inhibition of transcription complex formation by JAZ proteins. Methods Mol. Biol. 1011: 187–197. [DOI] [PubMed] [Google Scholar]
- Reeves P.H., et al. (2012). A regulatory network for coordinated flower maturation. PLoS Genet. 8: e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan S.M., Moffatt B.A. (1990). Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2: 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.-C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322. [Google Scholar]
- Sanders P.M., Lee P.Y., Biesgen C., Boone J.D., Beals T.P., Weiler E.W., Goldberg R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F., Fernández-Calvo P., Zander M., Diez-Diaz M., Fonseca S., Glauser G., Lewsey M.G., Ecker J.R., Solano R., Reymond P. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25: 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Huang H., Gao H., Wang J., Wu D., Liu X., Yang S., Zhai Q., Li C., Qi T., Xie D. (2014a). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Fan M., Zhang X., Gao H., Huang H., Wu D., Guo H., Xie D. (2013a). The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 9: e1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Xie D. (2013b). Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol. Plant 6: 1065–1073. [DOI] [PubMed] [Google Scholar]
- Song S., Qi T., Wasternack C., Xie D. (2014b). Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 21: 112–119. [DOI] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.Q., Jiang H.L., Li C.Y. (2011). Systemin/jasmonate-mediated systemic defense signaling in tomato. Mol. Plant 4: 607–615. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., Wang X.J., Hagen G., Guilfoyle T.J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Malek B., van der Graaff E., Schneitz K., Keller B. (2002). The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192. [DOI] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. (Lond.) 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xin X.F., He S.Y. (2013). Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51: 473–498. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V., Mallappa C., Gangappa S.N., Bhatia S., Chattopadhyay S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Christensen S., Isakeit T., Engelberth J., Meeley R., Hayward A., Emery R.J., Kolomiets M.V. (2012). Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell 24: 1420–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.Y., Li J.G., Pei M., Gu H., Chen Z.L., Qu L.J. (2007). Over-expression of a flower-specific transcription factor gene AtMYB24 causes aberrant anther development. Plant Cell Rep. 26: 219–228. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.