Summary
The intestinal microbiota, which is composed of bacteria, viruses, and micro-eukaryotes, acts as an accessory organ system with distinct functions along the intestinal tract that are critical for health. This review focuses on how the microbiota drives intestinal disease through alterations in microbial community architecture, disruption of the mucosal barrier, modulation of innate and adaptive immunity, and dysfunction of the enteric nervous system. Inflammatory bowel disease is used as a model system to understand these microbial-driven pathologies, but the knowledge gained in this space is extended to less well studied intestinal diseases that may also have an important microbial component, including environmental enteropathy and chronic colitis-associated colorectal cancer.
Introduction
The past decade has seen a dramatic rise in metagenomic and metabolomic studies of inflammatory bowel disease (IBD) and related inflammatory diseases. The most well understood IBDs include Crohn’s disease (CD) and ulcerative colitis (UC), which are chronic inflammatory disorders caused by multiple factors involving host genetics, the environment, and microbes. As a result, we are beginning to develop an ecological or community-wide understanding of the role of the microbiome in intestinal disease. Recently, these findings have begun to be translated into a functional mechanistic interpretation of the microbiome in inflammatory diseases.
In this review, we summarize the latest research on the role of the intestinal microbiome in inflammatory disease with a focus toward functional and mechanistic studies. We begin by exploring functional differences that exist along the length of intestinal tract and how these relate to IBD pathogenesis. We then introduce the major intestinal vulnerabilities that contribute to IBD and discuss functional evidence for how microbes contribute to either exacerbate or prevent onset of disease. Although most studies have focused on the bacterial component of the microbiome, we also discuss recent work that explores the impact of viral and microeukaryotic components. IBD can be considered the prototypic example of the potential for commensal microbes to influence intestinal disease, and here IBD is used as a context to interpret the role of the microbiome in other inflammatory bowel diseases, including environmental enteropathy, celiac disease, and colitis-associated colorectal cancer. Using this integrative approach, we highlight both recent advances in the field, as well as opportunities for novel therapeutic strategies for inflammatory bowel diseases.
Functional Differences Across the Intestinal Landscape
To appreciate how the microbiota influences chronic inflammatory diseases, especially IBD, it is critical to consider the physiological, immunological, and pathological differences along the intestinal tract. These aspects are often overlooked in studies concerned with defining the microbial changes that occur in IBD, yet they will be critical to understand the host pathways involved in disease initiation. Furthermore, many of the studies examining microbial changes in IBD have focused on stool samples, which are incompletely reflective of changes occurring at proximal sites of the intestine or in mucosal-associated communities. Functional differences in regions of the intestine are relevant, as CD can affect various areas of both the small and large intestine, resulting in a segmental pattern of inflammation. In contrast, UC tends to affect the colon, showing continuous inflammation. Here we will describe the major functional differences along the intestine in terms of the of composition of the epithelium, total microbial burdens, and secretion of antimicrobial peptides (AMPs) and mucus (Figure 1), focusing on how these functional differences are relevant to understanding and elucidating IBD pathogenesis.
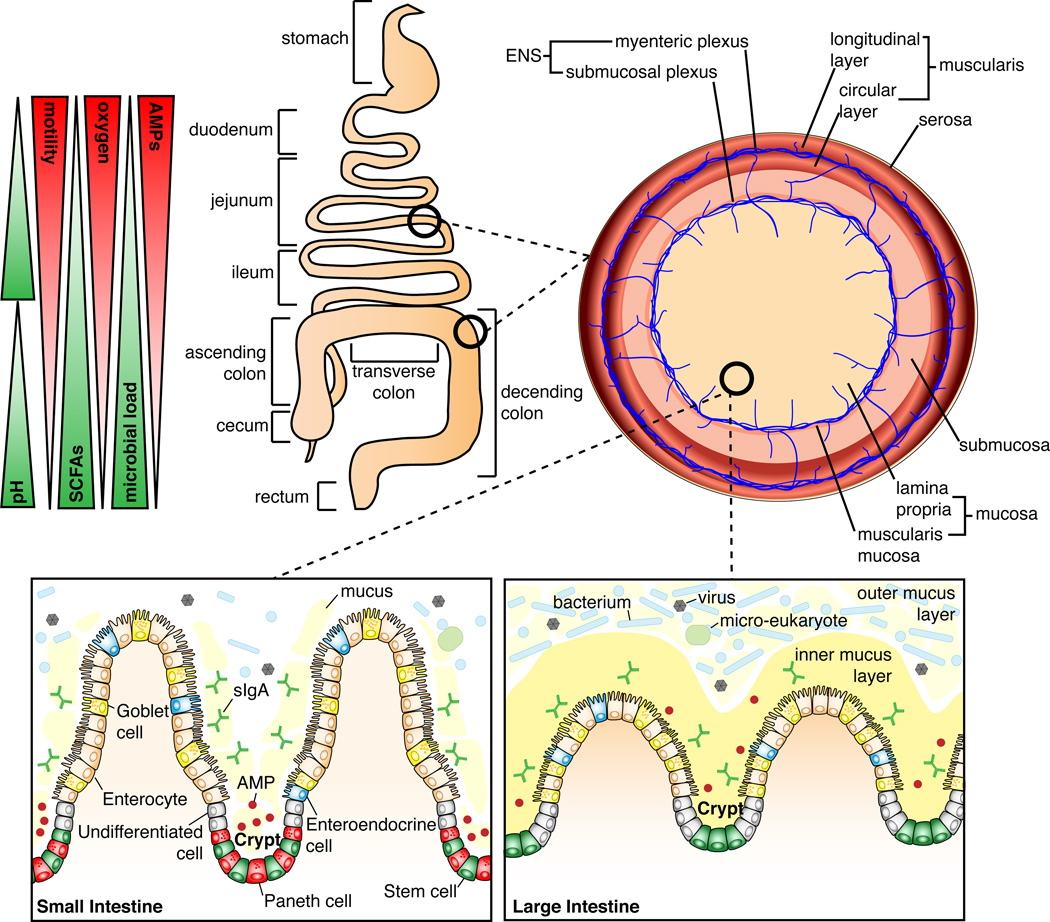
Figure 1. Differentiating features of the small and large intestinal landscape.
The small intestine begins after the stomach and is composed of the duodenum, jejunum, and ileum. The ileum joins to the large intestine via the cecum. The large intestine is composed of the ascending colon, transverse colon, descending colon, and rectum. The small intestine has higher oxygen levels and antimicrobial peptide (AMP) production, and increased intestinal motility, whereas in the large intestine, the microbial load is the highest and short-chain fatty acids (SCFAs) are abundant. The entire length of the intestine is lined by a single layer of epithelial cells. Below these cells is the lamina propria (LP), composed of connective tissue that provides the blood supply, lymphatic system, and enervation by the submucosal plexus, which are critical to the function of the intestine. Importantly, the LP houses many immune cells of both the innate and adaptive immune system (not shown). Further enteric enervation occurs in the thin layer of smooth muscle, the muscularis mucosa, which separates the LP from the underlying submucosa. Below the submucosa is a thick muscle layer, the muscularis, composed of an inner circular layer and outer longitudinal layer. Between the two muscle layers is the myenteric plexus, an important component of the enteric nervous system (ENS), which functions to coordinate intestinal peristalsis. The outermost covering of the intestine is the serosa. At the mucosal level the small intestine has long “finger-like” villi that project into the lumen, and which are absent in the large intestine. In the small intestine the crypts contain stem cells, AMP-producing Paneth cells, and undifferentiated cells; the villi contain the differentiated enterocytes, enteroendocrine cells, and goblet cells. In the small intestine goblet cells secrete mucus into the lumen, which has a loose, non-adherent, consistency. In the large intestine, the crypts lack Paneth cells and only contain stem cells and undifferentiated cells; the differentiated cells include enterocytes, enteroendocrine cells, and goblet cells. Here, enterocytes are involved in the production of AMPs and goblet cells secrete mucus that forms a bilayer structure: the inner and outer mucus layers. Although many of the cell types are shared between the small and large intestine, the function of these cells varies depending on the intestinal location.
Epithelial Layer
Although the small and large intestine have profound functional differences, they share some structural similarities (Figure 1). The small intestine is divided into three functionally distinct segments—the duodenum, jejunum, and ileum—and the function of the epithelium is regulated by the expression of transcription factors specific to each segment. For example, GATA4 is expressed by epithelial cells in the duodenum and jejunum, and reduction of GATA4 expression causes these cells to absorb bile acid, a function normally limited to epithelial cells of the ileum (Beuling et al., 2010). The majority of the digestive and absorptive function of the intestine occurs in the duodenum and jejunum, and is facilitated by long villi, as well as microvilli, which contain enzymes that mediate digestion and transport nutrients. One such brush border enzyme is intestinal alkaline phosphatase (IAP), which is highly expressed in the duodenum (Goldberg et al., 2008; Henthorn et al., 1987). IAP functions to hydrolyze monophosphate esters, resulting in detoxification of microbial ligands such as LPS, and is essential to maintain intestinal homeostasis (Bates et al., 2007; Goldberg et al., 2008). Inflamed mucosal tissue from intestines of patients with CD and UC exhibit reduced IAP production; this likely occurs through enhanced Toll-like receptor (TLR) 4 signaling and increased bacterial translocation into the mucosa (Goldberg et al., 2008; Molnár et al., 2012). Inflammatory diseases that affect the small intestine often result in blunting of villi, leading to malabsorption and malnutrition, as seen in celiac disease and environmental enteropathy (Dewar and Ciclitira, 2005; Kelly et al., 2004).
The main functions of the large intestine are the reabsorption of water and uptake of vitamins (e.g., vitamin K, vitamin B12, thiamine, riboflavin). The large intestine is also the site of enzymatic degradation of indigestible fiber by the microbiota, producing short-chain fatty acids (SCFAs). SCFAs, including acetate, propionate, and butyrate, exert a protective effect on epithelial cells and stimulate fluid absorption (Scheppach, 1994). It is well known that UC leads to changes in microbial composition and reduced production of SCFAs, and that treatment with SCFAs can be clinically beneficial (Harig et al., 1989). Mechanistically, butyrate is beneficial in UC as it is the major energy source for the epithelium, induces the differentiation of regulatory T cells (Tregs), and is critical in the resolution of inflammation by signaling through G-protein coupled receptor 43 (Furusawa et al., 2013; Maslowski et al., 2009).
Microbial Stratification Across the Intestine
The microbiota colonizes the entire length of the intestine with the total number of microbes increasing from the duodenum to the distal colon, where the microbial load is estimated to be 1012 microbes per gram of feces. There is a widespread belief that the duodenum and jejunum harbor less than 105 bacteria per gram of luminal content, and that this number increases in the ileum to up to 107 bacteria per gram of luminal content, with further increases in the colon (Berg, 1996; Mowat and Agace, 2014). A study by Vaishnava et al. (2011) showed that in the ileum of mice there are 107 mucosal-associated bacteria compared to 109 fecal bacteria, measured as 16S rRNA gene copies per cm2 of tissue or feces. Further, this study showed that a breakdown in the intestinal barrier mediated by a loss of MyD88 signaling in the epithelium resulted in a full log increase in mucosal-associated bacteria, with no change in fecal bacterial counts. Understanding how total bacterial load changes during IBD may be an interesting aspect of disease pathogenesis, as increased mucosa-associated bacteria could drive intestinal inflammation and may explain mixed patient responses to treatment.
The density of microbial colonization is mainly thought to be governed by pH, oxygen, and AMP gradients along the length of the intestine, as well as the speed of transit through the intestine (Figure 1). Reported pH values vary slightly but the trends are similar: pH is lowest in the duodenum (pH 6.4–6.6), gradually increases toward the terminal ileum (pH 7.3–7.5), decreases in the cecum (pH 5.7–6.4), and again gradually increases toward the distal colon (pH 6.6–7.1) (Evans et al., 1988; Fallingborg et al., 1989). An oxygen gradient exists along the length of the colon, with levels highest in the duodenum and decreasing to an anaerobic environment in the distal colon (He et al., 1999). The slightly acidic pH and oxygenated environment of the upper small intestine limits microbial colonization to acid- and oxygen-tolerant bacteria, mainly from the genera Lactobacillus, Streptococcus, and Veillonella (Frank et al., 2007; Zoetendal et al., 2012). There is also a radial oxygen gradient in the colon, with oxygen being produced by the epithelium and consumed by microorganisms living closest to the mucosa (Albenberg et al., 2014). The chronic inflammation associated with IBD likely leads to elevated oxygen levels in the intestine through increased blood flow and immunological responses such as the release of reactive oxygen species, which affect the microbial composition by selecting for more aerotolerant organisms and disrupting obligate anaerobic communities (Haberman et al., 2014; Rigottier-Gois, 2013). A recent study showed that treatment-naïve IBD patients at the time of diagnosis have an altered mucosal-associated bacterial community that favors the presence of aerotolerant species (Gevers et al., 2014; Haberman et al., 2014). This association of aerotolerant species could be used as an early diagnostic tool and suggests a novel therapeutic approach aimed at normalizing intestinal oxygen levels (Rigottier-Gois, 2013). Similar to the oxygen gradient, intestinal motility also decreases along the length of the intestinal tract, with the highest velocity documented in the duodenum and jejunum (Kellow et al., 1986). Changes in intestinal motility have an important impact on the severity of IBD (Bickelhaupt et al., 2013; Ohama et al., 2008), and a recent study showed that intestinal flares in CD are associated with a decrease in small intestinal motility and an increase in inflammatory markers such as C-reactive protein and calprotectin (Bickelhaupt et al., 2013).
Antimicrobial Peptides and the Mucus Layer
The small intestine has numerous Paneth cells located in crypts (Figure 1), which secrete AMPs including α-defensins, C-type lectins, lysozyme, and phospholipase A2 (Gallo and Hooper, 2012). AMP secretion is dependent on autophagy, as genetic disruption of autophagy stalls AMP secretion (Cadwell et al., 2008). In the large intestine, Paneth cells are absent, but AMP secretion, including the secretion of β-defensins, C-type lectins, cathelicidins, galectins, and lipocalin, is mediated by enterocytes (Gallo and Hooper, 2012). Interestingly, the microbiome of the large intestine encodes genes providing resistance to a specific class of AMPs (Cullen et al., 2015). This resistance confers resilience to the microbial community during inflammation, when AMP levels are high, allowing faster recovery of the microbiota after enteric infection (Cullen et al., 2015).
Intestinal mucus is produced by goblet cells and is mainly composed of a single, highly O-glycosylated protein, mucin 2 (MUC2), which is secreted into the lumen of the small and large intestine (Figure 1). Mucus-associated sugars are utilized by intestinal microbes and recent studies show that sensing of microbial ligands by type 3 innate lymphoid cells stimulates the addition of fucose to mucus, which in turn supports colonization of commensal bacteria (Goto et al., 2014; Pickard et al., 2014). The fucosidase and sialidase activity of certain symbiotic bacteria such as Bacteroides thetaiotaomicron to liberate mucosal glycans does not compromise the integrity of the mucus layer and is indeed a common feature of healthy microbiota. However, pathogenic bacteria can take advantage of these liberated sugars to facilitate their expansion (Ng et al., 2013). Further, establishing immunological tolerance to the intestinal microbiota has been shown to involve the exposure of dendritic cells to microbe-MUC2 complexes; recognition of which results in the generation of tolerogenic signals, dependent on the presence of MUC2, to specific microbial antigens (Shan et al., 2013). In the small intestine, microbe-triggered secretion of meprin β by enterocytes causes cleavage of MUC2 from the surface of the epithelium, preventing the formation of an adherent mucus layer (Schütte et al., 2014). In contrast, absence of meprin β in the large intestine allows the formation of a mucus bilayer (Schütte et al., 2014); the inner layer is firmly attached to the surface of the epithelium and is thought to be sterile, while the outer layer is a loose matrix containing microbes (Johansson et al., 2008). The physiological requirement for this bilayer structure is likely due to the high bacterial load of the large intestine, reduced AMP production, and the slower transit time of fecal matter. IBD patients have both a thinner inner mucus layer as well as reduced glycosylation of MUC2, with both alterations reducing the barrier potential of the inner mucus layer (Fyderek et al., 2009; Larsson et al., 2011). Like AMP secretion, mucus secretion in the large intestine is also highly dependent on autophagy, through signaling mediated by both reactive oxygen species and ligation of the NLRP6 inflammasome (Patel et al., 2013; Wlodarska et al., 2014).
Autophagy in two types of secretory cells in the intestine, Paneth cells and goblet cells, is highly relevant to IBD: a CD-associated risk allele in the autophagy gene ATG16L1 affects the function of both cell types (Hampe et al., 2007; Rioux et al., 2007). Genetic risk alleles could also contribute to disease by influencing composition of the microbiota; for example, monozygotic twins have a more similar intestinal microbiota than dizygotic twins (Goodrich et al., 2014), and IBD-related polymorphisms are associated with changes in abundance of specific bacterial taxa (Knights et al., 2014). The degree to which the functional properties of a host microbiota impact the bi-directional relationship between host genetics and microbiota colonization efficacy will be an important area of focus in the development of microbial therapeutics. For example, fecal microbiota transplantation (FMT) has proven efficacious in treating infectious colitis with C. difficile and, as we discuss below, there has been interest in treating IBD with FMT. However, it is unknown to what degree an altered genetic background in IBD patients could affect the therapeutic success.
Impact of the Intestinal Microbiome on the Vulnerabilities of IBD
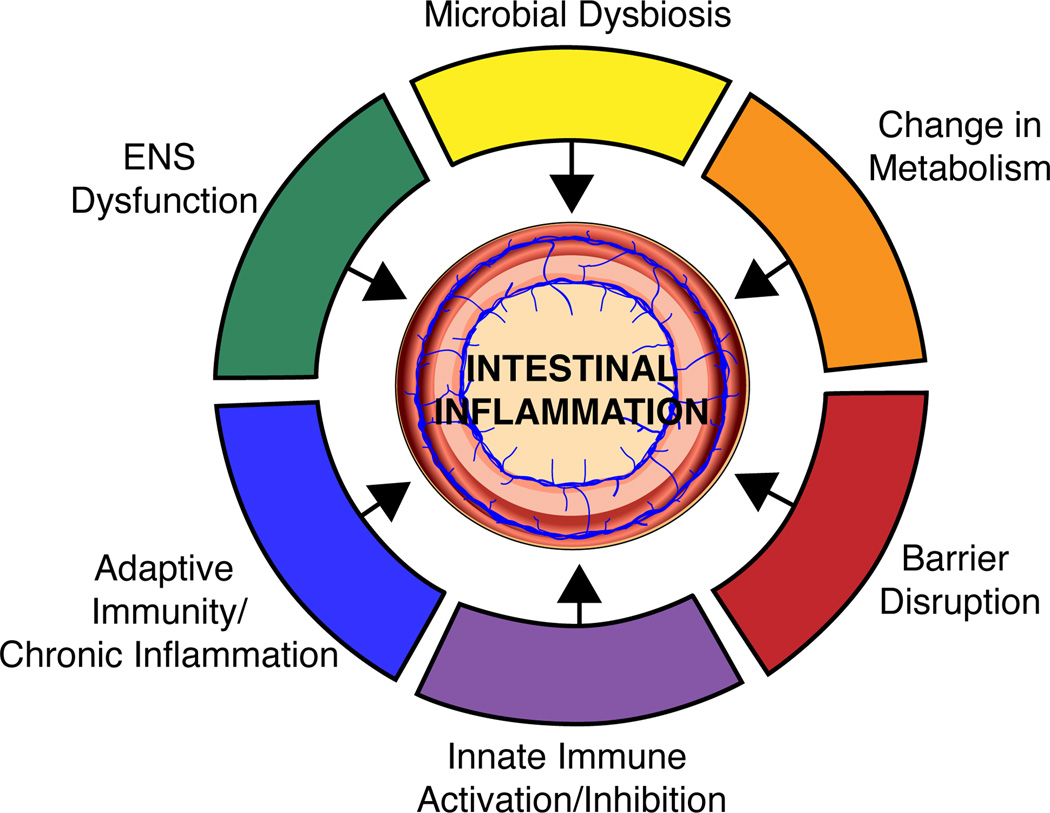
Metagenomic studies have so far led to a community-level understanding of recurrent alterations to the microbiome in IBD. Several groups have refined the resolution of this picture to identify specific mechanisms by which the intestinal microbiota affects the course of IBD and related inflammatory diseases. We attempt to organize these microbial-driven pathologies by introducing the major vulnerabilities of IBD below (Figure 2). As the microbiome research field matures to clinical translation in IBD, these vulnerabilities should serve as the primary targets of therapeutic intervention.
Figure 2. Intestinal vulnerabilities contributing to IBD.
Reduced alpha diversity and a change in the community architecture of the intestinal microbiota is a key phenotype of IBD and may contribute to disease initiation. The composition of the intestinal microbiota heavily influences the metabolic environment of the intestine and specific metabolites have been associated with inflammation. Further microbes influence the activity of both the innate and adaptive arms of the immune system and these interactions can initiate disease as well as maintain chronic inflammation. Intestinal microbes have been shown to produce a variety of neurotransmitters and are also critical for ENS development and function; these microbial-induced modifications to the ENS can translate to downstream effects on the mucosal immune system.
Microbial Dysbiosis
Recently, using a combined metagenomics- and host-transcriptome-based approach in a large cohort of treatment-naïve pediatric patients with CD, it has been possible to link specific alterations in the microbiome to host response in clinical disease. Gevers et al. identified a collection of bacteria that are differentially abundant in disease and demonstrated that this set of microbes hold predictive value for clinical outcomes in CD; the strongest signal was seen from biopsies of the terminal ileum, but surprisingly rectal biopsies were nearly as informative (Gevers et al., 2014). A subset of these patient biopsies were analyzed by host RNA-Seq, revealing a positive correlation between antimicrobial dual oxidase (DUOX2) expression with the expansion of Proteobacteria (found to be at increased abundance in CD), while expression of lipoprotein APOA1 was positively correlated with the abundance of the Firmicutes (decreased in CD). A regression model including both gene expression and microbial abundance accurately predicted remission of disease in this specific cohort of patients (Haberman et al., 2014). The signature of increased DUOX2 and decreased APOA1 expression favored oxidative stress and a Th1 phenotype, and was enriched in patients with severe mucosal injury (Haberman et al., 2014). Although these specific associations will need to be confirmed in expanded patient populations, this study provides an important framework for future studies integrating host expression and metagenomics datasets to make inferences into the interactions between host immune responses and the microbiome.
The infusion of feces from healthy donors, known as FMT, has been used for several decades to treat infection with Clostridium difficile, an endospore-forming, toxin-producing bacterium. However, the first controlled trial to demonstrate its efficacy was just recently published; this study showed that C. difficile-associated diarrhea was resolved after infusion of donor feces in 15 of 16 patients (van Nood et al., 2013). With this astounding success, FMT has become the new standard of care for C. difficile infection. In order to prevent the potential danger of infection from donor stool, Allen-Vercoe and colleagues, among other groups, have been designing synthetic mixtures of bacteria for stool transplant that have been successful in treating C. difficile infection (Petrof et al., 2013). Most recently, Buffie et al. (2014) used a workflow involving mouse models, clinical studies, metagenomic analysis, and mathematical modeling to identify a single species, the bile acid 7-alpha-dehydroxylating intestinal bacterium Clostridium scindens, that confers resistance to C. difficile infection and can be used to treat at-risk individuals.
IBD is objectively more complex than C. difficile infection because it is the dysbiotic community of bacteria in conjunction with aberrant mucosal immune responses that drive the patient toward disease. Several studies evaluating FMT for the treatment of IBD have been published to date, though the sample sizes are often small. One meta-analysis found that 80 of 113 (71%) of patients with IBD showed a reduction or resolution of symptoms upon treatment with FMT; however, it is unclear whether FMT will offer a curative response (Ianiro et al., 2014). A challenge in utilizing FMT for the treatment of IBD is the difficulty in predicting which donor strains will be maintained in the host and at what abundance; therefore, it is crucial to further investigate the factors that are important in species retention by the recipient and to develop retention prediction software for this purpose. In addition, the FMT approach for IBD should take into account both pre-treatment microbiome composition and immune phenotypes, allowing for the selection of a specific cocktail of microbes and immunomodulatory drugs to target the patient’s particular disease state.
Metabolic Effects
One mechanism by which the microbiome impacts inflammatory disease is by causing changes to the metabolism of the host. Host metabolism is deeply intertwined with that of the intestinal microbiome; for example, humans depend on symbiotic bacteria for the synthesis of several key vitamins including biotin, folate, vitamin B5, vitamin B6, riboflavin, and thiamin (Said, 2011). We also depend on our microbiome for a more complete extraction of nutrients from our diet beyond what we ourselves can digest. While the microbiome is an important contributor to metabolism in the maintenance of health, it can also push our metabolism toward inflammatory disease.
Hydrogen (H2) and hydrogen sulfide (H2S) production vary considerably across regions of the gastrointestinal tract and are involved in immunomodulation. In a study that examined the microbiome across 10 human body habitats (Segata et al., 2012), the authors found enzymes necessary for the production and utilization of H2, an anti-inflammatory metabolite (Kajiya et al., 2009) highly abundant in the oral cavity but nearly absent from the intestine. H2S is a metabolite produced by microbial H2 utilization, which at high concentrations can inhibit SCFA production and at low concentrations is involved in regulating host inflammatory responses (Segata et al., 2012). The potential for production of H2S is particularly high in stool and is strongly positively associated with the abundance of the pathogenic genera Treponema and Fusobacterium, suggesting a role in host-cell toxicity (Segata et al., 2012). A comparison of healthy human volunteers subjected to short term diets composed entirely of either plant or animal products, David et al. (2014) found that the microbiota of the carnivores contained more bile acid-tolerant taxa, including Alistipes, Bacteroides, and Bilophila, compared to vegans. High levels of Bilophila wadsworthia, which uses bile acids as an energy source, leads to increased taurine conjugation of bile acids and subsequent H2S production. High H2S levels in turn promote increased levels of proinflammatory Th1 cells and the development of colitis in IL-10−/− mice (Devkota et al., 2012). In pediatric patients with CD, an elemental diet consisting of amino acids, elemental nitrogen, carbohydrates, vitamins, minerals, and low fat has been shown to be as efficacious as corticosteroid therapy (Berni Canani et al., 2006). The elemental diet causes large changes in gut microbiome composition and increases microbial diversity (Shiga et al., 2012). As diet alters the availability of substrates for microbial generation of potentially harmful metabolites, this finding suggests the importance of diet in managing IBD.
Another important consideration in inflammatory disease is the impact of the microbiota on drug metabolism (Haiser et al., 2013; Maurice et al., 2013). For example, the cardiac glycoside digoxin causes the intestinal bacterium Eggerthella lenta to upregulate a unique operon encoding glycoside reductases that use digoxin as an electron acceptor, thus inactivating the drug (Haiser et al., 2013). The impact of the microbiome on the metabolism of drugs used to treat inflammatory and other diseases could help explain why certain drugs are more effective in some individuals than others and should be more closely studied.
Barrier Disruption
The separation of the microbiota from the host begins with the physical barriers: the mucus layer and the epithelium. Compartmentalization of the microbiota in the small intestine depends upon secreted antibacterial proteins, including RegIIIγ, α-defensins, and IgA (Gallo and Hooper, 2012). RegIIIγ is an antibacterial lectin that targets Gram-positive bacteria and is under the control of TLRs (Cash et al., 2006; Duerkop et al., 2009). Alpha-defensins are small antibacterial peptides which shape the composition of the luminal intestinal community (Salzman et al., 2003). In mice lacking an enzyme necessary to process mouse α-defensins, a large decrease in segmented filamentous bacteria and Th17 cells in the lamina propria was found, although total bacterial numbers were unchanged (Salzman et al., 2010). IgA is produced by plasma cells in response to antigens in the luminal environment, and the secreted form of IgA is transcytosed across the epithelium to allow binding to luminal bacteria (Macpherson and Uhr, 2004). Recently, Palm and de Zoete et al. exploited the tendency of IgA to target particularly pathogenic bacteria to identify a collection of colitogenic bacteria in human IBD that are heavily coated by IgA (Palm et al., 2014). These collections of bacteria were introduced into germ-free mice, which subsequently became more susceptible to chemically induced colitis (Palm et al., 2014). A new study observed striking changes in fecal IgA levels in wild-type mice that were vertically transmissible and correlated to susceptibility to chemically induced colitis, with lower IgA levels increasing susceptibility to colitis (Moon et al., 2015). Reduced IgA levels were shown to be due to an altered microbiome, characterized by the presence of a specific genus, Sutterella; this altered microbiome resulted in degradation of the secretory component of IgA (Moon et al., 2015). However, it remains to be determined which bacterial proteases are responsible for this degradation of the secretory component, as well as the relative abundance of these bacterial proteases in IBD patients.
The expression of antimicrobial peptides in the small intestine is controlled at least in part by mTOR, which is in turn is activated by the import of the amino acid tryptophan through the ACE2 transport pathway on the luminal surface of epithelial cells (Bröer et al., 2011; Farkas et al., 2006; Hashimoto et al., 2012; Reinisch et al., 2008). In the mouse, deficiency in Ace2, or a tryptophan-free diet, results in a drastically altered intestinal microbiome and increased severity of chemically-induced colitis. Transplantation of this altered microbiome to wild-type mice also results in the aggravation of colitis induced by dextran sodium sulfate (Hashimoto et al., 2012).
Innate Immunity
The mammalian immune system has co-evolved with its indigenous microbiota. It must maintain a rigid balance, on one hand exhibiting tolerance to symbiotic bacteria while on the other hand responding to threats from pathogenic bacteria. The breakdown of this balance lies at the core of most inflammatory diseases. NOD2, which is responsible for stimulating the immune system in response to bacterial peptidoglycans, was the first identified susceptibility gene for IBD (Hugot et al., 2001; Ogura et al., 2001) and is an example of a mechanism by which inflammatory disease is propagated by an overactive or underactive response to the symbiotic microbiota.
The microbiota delivers important cues to the host necessary for the maintenance of homeostasis and intestinal tolerance. Work in gnotobiotic zebrafish shows that the presence of the microbiota is essential for recruitment of neutrophils to sites of injury and found this effect to be mediated by the microbial-driven production of serum amyloid A (Kanther et al., 2014). Additionally, microbiota-driven IL-1β production by myeloid cells drives the maintenance of colonic regulatory Tregs in a process that requires production of granulocyte-macrophage colony-stimulating factor by type 3 innate lymphoid cells, resulting in Treg homeostasis and the production of regulatory factors including retinoic acid and IL-10 (Mortha et al., 2014). The presence of the symbiotic microbiota is necessary for a basal level of IL-1β production and the consequent homeostasis of several critical aspects of the immune system.
The intestinal epithelium expresses CD1d, which presents self and microbial lipid antigens to natural killer T (NKT) cells (Blumberg et al., 1991). CD1d is downregulated in inflamed IBD tissue, and NKT cells are involved in promoting colitis and IBD (Heller et al., 2002; Olszak et al., 2012). CD1d elicits its protective effects through the activation of STAT3 and the STAT3-dependent transcription of the regulatory cytokine IL-10, heat shock protein 110, and CD1d itself, and is an important mechanism by which the immune system maintains tolerance to its microbiota. Another recently discovered mechanism by which the microbiota tempers NKT-cell-mediated inflammation involves microbial production of sphingolipids, which are ubiquitous among eukaryotes but present in only a few bacterial genera. Wieland Brown et al. (2013) purified and solved the structures of three sphingolipids, also known as glycosylceramides, produced by a strain of Bacteroides. One of these, alpha-galactosylceramide, is capable of controlling NKT cell activity. Similarly, Kasper and colleagues have identified a glycosphingolipid produced by Bacteroides fragilis that inhibits invariant NKT (iNKT) cell activity, leading to a reduction in their numbers in the lamina propria and a decreased severity of disease in a mouse model of colitis (An et al., 2014). It is important to note that human studies are required to validate these findings as very few murine immune phenotypes have successfully been translated to human disease.
Adaptive Immunity
The function of the microbiome in adaptive immune cell homeostasis was first uncovered with the discovery that polysaccharide A produced by Bacteroides fragilis restores the Th1/Th2 balance in germ-free mice and protects from colitis by inducing Tregs (Mazmanian et al., 2005, 2008). Further, colonization of mice with segmented filamentous bacteria (SFB) in the small intestine results in the adherence of SFB to the absorptive cells of the ileum and those over Peyer’s patches, promoting the production of Th17 cells, which are the sole subset of T cells in the intestine able to detect SFB antigens (Ivanov et al., 2008; Schnupf et al., 2015; Yang et al., 2014). Since these initial studies, Honda and colleagues have shown that a group of 46 murine-derived and 17 human-derived Clostridium species can induce colonic Tregs in mice, resulting in protection from colitis and allergy (Atarashi et al., 2013). However, there is no effect on Tregs when any one of these species is used to mono-colonize a germ-free mouse, suggesting that some aspect of the community-level interactions is required to induce Tregs rather than any single Clostridium species (Atarashi et al., 2013). The SCFAs also play an important role in the maintenance of host adaptive immune function. SFCAs contribute to protection from inflammation (Maslowski et al., 2009) and induce Treg function by inhibiting histone deacetylase activity, leading to an increase in acetylation of the Foxp3 gene and increased Foxp3 expression (Arpaia et al., 2013; Atarashi et al., 2013; Furusawa et al., 2013; Smith et al., 2013b). Future studies should focus on validating these pathways in human gut mucosa as the effect of these specific microbes on the adaptive immune system of mice will not necessarily translate to human disease.
Enteric Nervous System
The enteric nervous system (ENS) is made up of 200–600 million neurons that are organized into two plexuses: the myenteric plexus, which extends the entire length of the gastrointestinal tract, and the submucosal plexus, only found in the small and large intestine (Figure 1). The main functions of the ENS include regulation of intestinal peristalsis, transmucosal fluid flux, local blood flow, release of intestinal hormones, nutrient absorption, and interactions with the immune system. The effect of the intestinal microbiota on the ENS is an emerging area of research and in-depth characterization of enteric neurons, enteric glial cells (EGCs), and enteroendocrine cells is required to understand how microbes communicate with these cells, as well as the downstream effects on intestinal immunity and physiology, and how this impacts IBD.
Several molecules with neuroactive function, including gamma-aminobutyric acid (GABA), tryptamine, indole-3-propionic acid, serotonin, and SCFAs, are produced by intestinal bacteria and act on the ENS to modulate intestinal homeostasis (Table 1). GABA is a well-known enteric neurotransmitter found at high levels in the myenteric plexus and endocrine cells in the intestine, and is an important modulator of intestinal motility (Krantis, 2000). Interestingly, GABA signaling in the CNS is linked to anxiety and depression, which are often associated with IBD. In mice, ingestion of a Lactobacillus rhamnosus strain results in altered GABA expression in the brain; however, it is unclear how GABA signaling in the ENS contributes to this observation (Bravo et al., 2011). GABA is produced by the host using glutamate as a substrate in a reaction involving L-glutamate decarboxylase, and it was recently shown that certain strains of Lactobacillus and Bifidobacterium also produce GABA in the intestine using the same biosynthetic pathway (Barrett et al., 2012). The production of GABA by the intestinal microbiome is also affected by diet: in carnivores, catabolic reactions to break down glutamate are increased (potentially leading to an increase in intraluminal GABA) whereas herbivores show an increase in glutamate biosynthesis pathways (Muegge et al., 2011). The amino acid tryptophan can also be utilized by the intestinal microbiota, and metabolic byproducts include the production of tryptamine, serotonin, and indole-3-propionic acid (Wikoff et al., 2009; Williams et al., 2014). Both tryptamine and serotonin are neurotransmitters with documented effects on intestinal motility (Fayyaz and Lackner, 2008; Williams et al., 2014), whereas the function of indole-3-propionic acid is less understood, but has been found in cerebrospinal fluid and has a neuroprotective function (Chyan et al., 1999). Further, serotonin produced by enteroendocrine cells in the intestine has been shown to contribute to inflammation during chemically-induced colitis in mice, and inhibiting the production of mucosal serotonin is sufficient to reduce inflammation (Margolis et al., 2014). How microbial production of serotonin contributes to intestinal inflammation during IBD is not well described. Microbial produced SCFAs have also been shown to enhance enteric neuronal maturation and intestinal motility (Suply et al., 2012).
Table 1.
Neuroactive molecules produced by commensal bacteria in the intestine.
| Neuroactive Molecules |
Bacterium | Catalytic enzyme/process | Effect | Ref |
|---|---|---|---|---|
| GABA |
Lactobacillus, Bifidobacterium |
Glutamate decarboxylase/ Decarboxylation of glutamate to GABA |
Prevents anxiety and depression Regulates intestinal motility |
(Barrett et al., 2012; Bravo et al., 2011; Krantis, 2000) |
| Tryptamine |
Clostridium sprogenes, Ruminococcus gnavus |
Trp decarboxylase/ Decarboxylation of tryptophan to tryptamine |
Tryptamine induces ion secretion by IECs Tryptamine induces the release of serotonin |
(Williams et al., 2014) |
|
Indole-3- Propionic Acid |
Clostridium sporogenes |
Unknown/ Deamination of Tryptophan |
Powerful antioxidant Production requires microbiota |
(Chyan et al., 1999; Wikoff et al., 2009) |
| Serotonin | Unknown | Unknown/ Metabolite of Tryptophan |
Stabilizes mood/ anti-depressant Modulates intestinal motility |
(Fayyaz and Lackner, 2008; Wikoff et al., 2009) |
| SCFA |
Bacteroides, Bifidobacterium, Propionibacterium, Eubacterium, Lactobacillus, Clostridium |
Multiple/ Fiber fermentation | Energy source Reduce proinflammatory cytokine production |
(Macfarlane and Macfarlane, 2012; Suply et al., 2012) |
EGCs, the major cellular component of the ENS, are found in the mucosa and are essential to ENS function and intestinal homeostasis. EGCs are closely organized with enteric neuronal structures, and maintenance of EGC numbers in the mucosa is dependent on presence of the microbiota (Kabouridis et al., 2015). Ablation of EGCs results in severe intestinal inflammation due to a defective intestinal barrier and has been proposed as one mechanism of CD initiation, as early pathological changes to EGCs has been reported in patients (Cornet et al., 2001). EGCs express TLRs 2, 3, and 4, and detection of bacterial microbe-associated molecular patterns by EGCs has a direct effect on the barrier function of the intestinal epithelium (Savidge et al., 2007). Increased susceptibility of Tlr2-deficient mice to chemically induced colitis has recently been shown to result from alterations in the architecture of the ENS and EGC function (Brun et al., 2013). Intestinal TLR signaling in response to LPS is also critical in peristalsis by directing the production of CSF-1 by enteric neurons. This signaling results in macrophage recruitment to the intestinal muscularis and production of bone morphogenic protein 2, which acts on enteric neurons that direct peristalsis (Muller et al., 2014). During antibiotic use, this TLR signaling was lost, with severe effects on peristalsis. The addition of LPS was able to reverse this defect, but it remains unclear which cell types propagate the LPS signal to the enteric neurons (Muller et al., 2014).
IBD and the Neglected Microbes: Micro-Eukaryotes and Viruses
The intestinal microbiome harbors a micro-eukaryote and viral community that has been mostly overlooked to date. Here we highlight recent studies that are beginning to explore the importance of these microbial communities and their relevance to IBD.
Micro-eukaryotes
Similar to the bacterial component of the human fecal microbiota, the micro-eukaryote community is dependent on geography (Parfrey et al., 2014; Yatsunenko et al., 2012). The fecal microbiome of United States residents, as compared to Malawians, has reduced abundance of fungi and other micro-eukaryotes both in adult and child populations (Parfrey et al., 2014; Yatsunenko et al., 2012). This shift in the abundance of intestinal micro-eukaryotes is likely to be relevant to the hygiene hypothesis, which describes the association of increasingly hygienic conditions and allergic disease, and may be a factor in the increased prevalence of IBD in developed countries.
Generally, the abundance of microbes is determined as a percentage of total sequencing reads, with the micro-eukaryotes consisting of 0.01–0.1%, suggesting low abundance (Parfrey et al., 2014). However, this methodology likely under-represents the total micro-eukaryote community, as it does not consider the microbiome composition in terms of total biomass (with micro-eukaryotes such as fungi outweighing a typical bacterial cell by 100X). In addition, the identified sequencing reads require annotated reference sequences that are not well developed for microeukaryotes. Furthermore, no studies have examined the presence of micro-eukaryotes in the mucosal-associated portion of the human intestinal microbiota, as all studies have relied on fecal-derived DNA for sequencing. From a metabolic perspective, the micro-eukaryotes have a greater potential for generating a larger small-molecule output compared to bacteria.
A common trend emerging from genomic studies that probe the microeukaryotic population is the widespread prevalence of the intestinal protist, Blastocystis, in healthy humans around the world (Alfellani et al., 2013; Nam et al., 2008; Scanlan et al., 2014). The vast heterogeneity between the nine known Blastocystis subtypes that colonize humans, as well as their low level abundance, has led to wide variation in reports on the prevalence of colonization: 5–56% of individuals in developed countries and up to 76% in developing countries (Tan, 2008; Turkeltaub et al., 2015). In the limited studies that have directly assessed the possible role of micro-eukaryotes in IBD, Petersen et al. showed that in Danish cohorts, colonization by microeukaryotes, including Blastocystis and Dientamoeba, is negatively associated with active UC, suggesting a protective nature of these protists (Petersen et al., 2013). Although it is unlikely that a lack of a specific microeukaryote is responsible for onset of IBD, they may have important metabolic functions that reduce susceptibility to chronic intestinal inflammation.
The human fecal microbiome includes a rich fungal community. This observation has also been made for the murine microbiome, allowing mechanistic insight into the role of fungi in intestinal health (Iliev et al., 2012; Scupham et al., 2006). Dectin-1 is a C-type lectin expressed on the surface of neutrophils, macrophages, and dendritic cells, which recognizes β-1,3-glucans found in the cell wall of fungi; ligation of dectin-1 initiates a Th17-mediated immune response. Iliev and colleagues (2012) showed that fungi are closely associated with the mucosa and that fungal-dependent signaling through dectin-1 is important for maintaining tolerance to the microbiota and protection from intestinal inflammation. In line with this hypothesis, a recent study showed that the association of mucus with commensal bacteria initiates a tolerogenic signal to dendritic cells of the small intestine that requires signaling through dectin-1 (Shan et al., 2013). Further, a mutation in human dectin-1 is associated with medically refractory UC (Iliev et al., 2012). Aside from the study by Iliev et al., very little is known regarding how fungi contribute to host metabolism or inflammatory pathways in IBD. Sphingolipid signaling in the host controls many aspects of inflammatory diseases, and it is now appreciated that not all sphingolipids are host-derived (Maceyka and Spiegel, 2014). While only a small subset of intestinal bacteria produce sphingolipids, all fungi contain sphingolipids in their membranes (Olsen and Jantzen, 2001), with yeast being the best characterized example (Ejsing et al., 2009). One study showed that yeast-derived sphingolipids can activate host peroxisome proliferator-activated receptors in the liver (Murakami et al., 2011); however, an understanding of how these sphingolipids integrate into host inflammatory pathways is lacking. These studies will require well-controlled germ-free transfer studies of micro-eukaryote isolates, and better determination of the relative lipidomic profiles of micro-eukaryotic species and how they interact with the mucosal immune system of the intestine.
Viruses
A second neglected aspect of the microbiome is the symbiotic virome. Although viruses have historically been considered only in the context of single disease-single pathogen relationships, advances in sequencing technology have uncovered the dramatic size of the virome present in health and disease: it is estimated that there may be 10-fold more viruses in the human microbiome than there are bacteria (Mokili et al., 2012), meaning the virome might outnumber human cells by 100-fold (Breitbart et al., 2003; Minot et al., 2011, 2012, 2013; Reyes et al., 2010). The human virome is composed of both eukaryotic viruses as well as bacteriophages, which directly affect the bacterial microbiome composition. For example, Rohwer and colleagues found that phage capsids express an immunoglobulin-like protein domain that binds to mucin glycoproteins and may be a source of non-host-derived innate immunity (Barr et al., 2013). Therefore, a large ecosystem of viruses exists in the healthy state “asymptomatically,” including many viruses that have previously only been considered in the context of pathogenicity. In recent years there has been a wealth of data emerging that describes manifold roles for the virome in inflammatory disease and in maintaining health. In a recent study, Virgin and colleagues found that some changes in the composition of the bacterial community in IBD may be explained by shifts in the bacteriophage community (Norman et al., 2015). Using a longitudinal study design of patients with UC and CD paired with household healthy controls, the authors described a significant expansion of the Caudovirales bacteriophages in both CD and UC, and a generalized increase in diversity of the virome concomitant with a decreased bacterial microbiome diversity (Norman et al., 2015). Though it is possible that the expanded Caudovirales may be directly responsible for decreased bacterial diversity, it is not clear whether the increased abundance of the bacteriophage is acquired from the environment or caused by an induction of prophage by the symbiotic microbiota. Kernbauer et al. (2014) reported that colonizing germ-free mice with a single virus, namely murine norovirus, can replace many of the beneficial functions of commensal bacteria. Further, the virus leads to improvement of several other deficiencies observed in germ-free mice; these benefits include an increased number of CD3+ T cells, increased Paneth cell function, and higher antibody levels. The ability of both bacteria and a single virus to re-model the germ-free intestine suggests that this effect largely occurs through non-specific microbial signals. Furthermore, in response to dextran sodium sulfate exposure, murine norovirus improves overall survival and prevents colon-length shortening (Kernbauer et al., 2014).
Although the interplay between genetics and the microbiome has been explored largely in terms of the bacterial component, recent studies also point to the importance of viruses. In mice with reduced expression of the autophagy gene Atg16l1, Paneth cells display a number of structural abnormalities and a dysfunction in granule secretion (Cadwell et al., 2008). However, these mice appeared normal under germ-free conditions, and introduction of an intestinal norovirus was sufficient to restore the Paneth cell dysfunction (Cadwell et al., 2010). Similarly, infection of IL-10-deficient mice with norovirus resulted in epithelial barrier disruption, illustrating that norovirus provides a colitogenic stimulus that can push the host toward colitis depending on the composition of the remainder of the intestinal microbiota (Basic et al., 2014). These examples demonstrate the significance of viruses in mediating disease phenotypes in genetically susceptible hosts.
IBD as a Platform to Understand other Intestinal Diseases
Aside from IBD, there are many other prevalent intestinal inflammatory diseases that have not been well explored in terms of the impact of the microbiome. The knowledge gained from mining the intestinal microbiome and defining the microbial-driven pathologies contributing to IBD can now be applied to these understudied illnesses to unravel their etiology and develop novel therapeutics.
The Intestinal Microbiome, Inflammation, and Global Health
Aside from CD and UC, which generally impact individuals living in high-income countries, distinct IBD-like syndromes exist that are far more prevalent in low-income countries where poor sanitation, hygiene, and malnutrition are widespread. These include environmental enteropathy (EE), tropical sprue (TS), and immunoproliferative small intestinal disease (IPSID). EE and TS are now recognized to be significant contributors to the sequelae and persistence of childhood malnutrition worldwide, which is the leading cause of childhood mortality (Korpe and Petri, 2012). IPSID affects young adults who suffer from chronic inflammation of the small intestine, resulting in lymphoma. However, despite the wide impact of these diseases, their exact cause(s) remain unknown. Poor sanitation in low-income countries could lead to higher exposure to fecal bacteria and increased incidence of enteric infection. The continuous exposure to fecal bacteria could result in bacterial overgrowth in the small intestine; numerous studies have found such overgrowth in children living with EE symptoms, by utilizing hydrogen breath tests (Fagundes Neto et al., 1994; Khin-Maung-U et al., 1992; dos Reis et al., 2007) or direct sampling of the small intestinal aspirate (Heyworth and Brown, 1975; Omoike and Abiodun, 1989). This overgrowth of microbes may cause the pathological changes characteristic of EE, in particular, villous blunting, increased intestinal permeability, and chronic inflammation that impair the proximal small intestine and subsequently reduce nutrient absorption (Lin et al., 2013; Ngure et al., 2014; Weisz et al., 2012). The transfer of fecal microbiota from severely malnourished children who exhibit EE features was shown to recapitulate EE disease in the small intestine of germ-free mice, including villous blunting (Smith et al., 2013a). Eleven strains of IgA-coated bacteria, including Enterobacteriaceae, were isolated from the fecal microbiota of these children, and were shown to confer the EE-like phenotype in germ-free mice, suggesting that these strains may be responsible for disease pathology (Kau et al., 2015). Alternatively, enteric infection can drastically remodel the intestine, making it more susceptible to chronic disease that would be exacerbated with poor sanitary conditions. The contribution of these microbial changes to the pathophysiology of EE, TS, and IPSID remain mostly unknown and, similar to IBD, there is no one pathogen, microbe, gene, or environmental factor that has been shown to drive disease.
The definition of EE and TS as distinct entities is a source of debate; however, consensus exists on some important defining features. EE and TS are similar in that both diseases cause growth stunting and have similar hallmarks, including chronic inflammation, malabsorption, villous atrophy, lymphocyte infiltration, and increased permeability in the small intestine (Keusch et al., 2014). However, they differ as EE does not clinically present with diarrhea whereas TS presents with chronic diarrhea and is infectious in nature (Korpe and Petri, 2012). An inadequate diet impacts the immune system, increasing the incidence and severity of infection in TS, which leads to an altered metabolism and nutrient loss, exacerbating malnutrition (Katona and Katona-Apte, 2008; Mondal et al., 2012; Schaible and Kaufmann, 2007). If the enteric infection is cleared in TS, it is not known whether disease would persist in malnourished children in the form of post-infectious EE and hence it is unclear if these two diseases are independent of each other.
IPSID is a form of small intestinal lymphoma that is mainly seen in developing countries (Al-Saleem and Al-Mondhiry, 2005). This type of lymphoma arises from lymphoid infiltration of the mucosa-associated lymphoid tissue, leading to malabsorption and protein-losing enteropathy; in advanced stages of disease, a malignant diffuse large-B cell lymphoma will form (Al-Saleem and Al-Mondhiry, 2005). From a microbiological perspective, this unique small intestinal cancer is particularly interesting: early stages of disease, prior to tumor development, are responsive to antibiotics, strongly suggesting that the disease may be triggered by microbial infection or microbial dysbiosis. No single bacterial pathogen has been identified as the cause of IPSID, although Lecuit et al. (2004) reported that 71% of patients examined had Campylobacter jejuni associated with the jejunal mucosa, and a beneficial response to antibiotic treatment was associated with eradication of C. jejuni. However, changes to other members of the microbiome have not been explored.
The Intestinal Microbiota, Chronic Inflammation, and Colorectal Cancer
Several recent studies implicate the intestinal microbiota as an important player in colorectal cancer (CRC) (Irrazábal et al., 2014; Kostic et al., 2013a; Sears and Garrett, 2014). Some of the alterations observed in the CRC microbiome are also observed in IBD, and chronic intestinal inflammation is a major risk factor for the development of CRC. Human studies comparing bacterial communities in tumors relative to noninvolved adjacent colonic tissue, have identified Fusobacterium species as the most strongly enriched taxon at the tumor site (Castellarin et al., 2012; Kostic et al., 2012; Marchesi et al., 2011). Fusobacterium is a colonocyte-invading bacterium also implicated in IBD (Strauss et al., 2011); as IBD is one of the greatest risk factors for developing CRC, this inflammation-associated bacterium may prove to be an important link between these diseases. More recent studies have associated Fusobacterium with colorectal adenomas (Kostic et al., 2013b; McCoy et al., 2013), the precursors to CRC, lending evidence that Fusobacterium is present, if not directly involved, in the tumorigenic process. The introduction of human colonic isolates of Fusobacterium nucleatum to genetically susceptible mice resulted in the acceleration of intestinal tumorigenesis (Kostic et al., 2013b; Rubinstein et al., 2013), further lending evidence to a direct role of Fusobacterium in driving cancer.
Other Intestinal Inflammatory Disease States
Celiac disease, Prevotella-driven disease, and type 1 diabetes (T1D) have all been associated with particular microbial signatures. Celiac disease is an immune-mediated enteropathy triggered by dietary wheat gluten in genetically susceptible individuals. The HLA-DQ genes are the major genetic determinant of disease, accounting for 40% of the genetic risk. However, a decreased abundance of Bifidobacterium and increased numbers of Staphylococcus and Bacteroides fragilis were also shown in infants at higher genetic risk for celiac disease and may contribute to disease progression (Palma et al., 2012). Prevotella, an emerging pathobiont implicated in inflammatory and autoimmune diseases, has been shown to invade colonic crypts, drive NLRP6-mediated dysbiosis, and exacerbate colitis (Elinav et al., 2011). A stool-based study of 114 new-onset untreated rheumatoid arthritis patients and controls identified Prevotella copri as being strongly associated with disease, and colonization of mice with P. copri in resulted in more severe chemically induced colitis (Scher et al., 2013). Interestingly, Pstpip2cmo mice, which develop spontaneous osteomyelitis, have an intestinal microbiota that is characterized by outgrowth of Prevotella species (Lukens et al., 2014). A recent prospective longitudinal study of infants at risk for T1D identified a microbial signature consisting of a steep drop in community diversity and an expansion of pathobionts including the Rikenellaceae. This signature correlated with higher fecal β-defensin 2 levels, and differentiated patients who went on to develop T1D versus non-T1D individuals one year before clinical diagnosis (Kostic et al., 2015). This study suggests, as has been shown previously in mice, that a pro-inflammatory intestinal microbiota may promote extra-intestinal inflammatory and autoimmune disorders such as T1D (Markle et al., 2013; Wen et al., 2008; Yurkovetskiy et al., 2013). Further longitudinal studies that investigate the microbiome during human disease are required to clarify whether these changes are a cause or a consequence of these and other diseases.
Conclusion
Early investigators of IBD searched for a single pathogenic microbe or pathway responsible for disease; however, recent studies have proved the inherent flaw in this approach. Relationships between microbes and host processes in the intestine represent a complex, adaptive system, constantly in flux. There is incredible inter- and intra-person variability of the microbiome with less than 10% of our microbes shared across individuals. Despite this complexity, there are some fingerprints seen in intestinal disease that are shared, such as the observed overall reduction in microbial diversity and increased abundance of mucosal-associated aerotolerant bacteria. The reported microbial changes in IBD patients can be considered a microbial fingerprint that represents an average across an entire cohort of IBD patients. Although such a fingerprint is useful, what is needed to enhance our understanding of IBD is stratification of IBD patients based on specific types of microbial shifts and the development of a classification system for these microbial fingerprints. With such a system, it may be possible to understand whether specific microbial fingerprints are associated with specific patient genetic mutations and whether specific microbial fingerprints can be used to predict patient response to current therapeutics. Identification of the relationships between genetics and microbes are required in order to understand how these associations can be utilized to logically target specific treatments for CD and UC, as well as other intestinal disease. For example, in an IBD patient with a NOD2 polymorphism, which supports the expansion of Enterobacteriaceae (Knights et al., 2014), perhaps the ideal synthetic microbial community would involve beneficial Clostridium strains to enhance Treg function, in combination with antimicrobials to selectively eliminate strains within the Enterobacteriaceae family.
IBD can affect multiple areas of the intestinal tract, which varies greatly along its length with regard to its associated bacterial community, and immunological and physiological functions. Therefore, it is likely that the pattern of inflammation and microbial changes within a patient is critical in understanding the trigger of disease. This is a largely unexplored area and would require characterization and stratification of IBD patients beyond the classical CD and UC classification scheme. Critical to moving toward patient stratification is the ability to assess the small intestinal microbiome during disease in patients without requiring patient bowel preparation (enema or laxative), as this likely changes the mucosal-associated microbiome and may mask important microbial phenotypes. This goal could potentially be achieved with the generation of ingestible nanosensors to detect microbes and other physiologically relevant measures of intestinal health (e.g., oxygen levels, pH, gastrointestinal motility). Of interest to the spatial distribution of IBD, treatment-naïve CD patients show significant changes in mucosal-associated bacteria that are similar in the ileum and the rectum (Gevers et al., 2014). However, if the microbial changes are similar in both sites, why does CD occur only in the ileum and not progress to involve large intestinal inflammation? If this change in bacterial community structure is driving disease, what are the key interactions between bacteria in the ileum and the mucosa that are absent in the rectum?
In addition to the pharmaceutical interest in generating synthetic microbial communities, further understanding of the metabolites and other compounds produced by our microbiome (bacterial, viral, and micro-eukaryotic) is another interesting avenue to develop new therapeutics for IBD and other chronic inflammatory conditions. The metabolites produced by members of our intestinal microbiota have largely been unexplored. However, the application of metabolomic and metatranscriptomic approaches to IBD patient cohorts may point to bacterial products that are overabundant or absent in IBD that may have therapeutic potential. Lastly, there is a great need for hypothesis-based mechanistic studies that utilize what has been learned through sequencing efforts to focus on understanding the many uncharacterized microbe-host and microbe-microbe interactions that are critical for maintaining intestinal health. By understanding these uncharacterized pathways, we may begin to understand the interplay of genetics, microbes, and environment contributing to pathogenesis of intestinal diseases.
The human gut microbiota plays an important role in maintaining a healthy gastrointestinal tract. In this review, Wlodarska et al. discuss the current knowledge of host-microbiota interactions in gut health, and how microbial dysbiosis contributes to the development and pathology of inflammatory diseases of the intestine, including Inflammatory Bowel Disease.
Acknowledgements
A.D.K. is supported by a Helen Hay Whitney Foundation Research Fellowship and the Lawrence H. Summers Postdoctoral Fellowship. R.J.X. is supported by grants from the Crohn’s and Colitis Foundation of America, the Leona M. and Harry B. Helmsley Charitable Trust, JDRF, and the National Institutes of Health. We thank Eric M. Brown and Natalia Nedelsky for thorough editing of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Saleem T, Al-Mondhiry H. Immunoproliferative small intestinal disease (IPSID): a model for mature B-cell neoplasms. Blood. 2005;105:2274–2280. doi: 10.1182/blood-2004-07-2755. [DOI] [PubMed] [Google Scholar]
- Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfellani MA, Stensvold CR, Vidal-Lapiedra A, Onuoha ESU, Fagbenro-Beyioku AF, Clark CG. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013;126:11–18. doi: 10.1016/j.actatropica.2012.12.011. [DOI] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E, Ross RP, O’Toole PW, Fitzgerald GF, Stanton C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012;113:411–417. doi: 10.1111/j.1365-2672.2012.05344.x. [DOI] [PubMed] [Google Scholar]
- Basic M, Keubler LM, Buettner M, Achard M, Breves G, Schröder B, Smoczek A, Jörns A, Wedekind D, Zschemisch NH, et al. Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm. Bowel Dis. 2014;20:431–443. doi: 10.1097/01.MIB.0000441346.86827.ed. [DOI] [PubMed] [Google Scholar]
- Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- Berni Canani R, Terrin G, Borrelli O, Romano MT, Manguso F, Coruzzo A, D’Armiento F, Romeo EF, Cucchiara S. Short- and long-term therapeutic efficacy of nutritional therapy and corticosteroids in paediatric Crohn’s disease. Dig. Liver Dis. 2006;38:381–387. doi: 10.1016/j.dld.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Beuling E, Kerkhof IM, Nicksa GA, Giuffrida MJ, Haywood J, aan de Kerk DJ, Piaseckyj CM, Pu WT, Buchmiller TL, Dawson PA, et al. Conditional Gata4 deletion in mice induces bile acid absorption in the proximal small intestine. Gut. 2010;59:888–895. doi: 10.1136/gut.2009.204990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickelhaupt S, Pazahr S, Chuck N, Blume I, Froehlich JM, Cattin R, Raible S, Bouquet H, Bill U, Rogler G, et al. Crohn’s disease: small bowel motility impairment correlates with inflammatory-related markers C-reactive protein and calprotectin. Neurogastroenterol. Motil. 2013;25:467–473. doi: 10.1111/nmo.12088. [DOI] [PubMed] [Google Scholar]
- Blumberg RS, Terhorst C, Bleicher P, McDermott FV, Allan CH, Landau SB, Trier JS, Balk SP. Expression of a nonpolymorphic MHC class I-like molecule, CD1D, by human intestinal epithelial cells. J. Immunol. 1991;147:2518–2524. [PubMed] [Google Scholar]
- Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröer A, Juelich T, Vanslambrouck JM, Tietze N, Solomon PS, Holst J, Bailey CG, Rasko JEJ, Bröer S. Impaired nutrient signaling and body weight control in a Na+ neutral amino acid cotransporter (Slc6a19)-deficient mouse. J. Biol. Chem. 2011;286:26638–26651. doi: 10.1074/jbc.M111.241323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun P, Giron MC, Qesari M, Porzionato A, Caputi V, Zoppellaro C, Banzato S, Grillo AR, Spagnol L, De Caro R, et al. Toll-like receptor 2 regulates intestinal inflammation by controlling integrity of the enteric nervous system. Gastroenterology. 2013;145:1323–1333. doi: 10.1053/j.gastro.2013.08.047. [DOI] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2014;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyan YJ, Poeggeler B, Omar RA, Chain DG, Frangione B, Ghiso J, Pappolla MA. Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid. J. Biol. Chem. 1999;274:21937–21942. doi: 10.1074/jbc.274.31.21937. [DOI] [PubMed] [Google Scholar]
- Cornet A, Savidge TC, Cabarrocas J, Deng WL, Colombel JF, Lassmann H, Desreumaux P, Liblau RS. Enterocolitis induced by autoimmune targeting of enteric glial cells: a possible mechanism in Crohn’s disease? Proc. Natl. Acad. Sci. U. S. A. 2001;98:13306–13311. doi: 10.1073/pnas.231474098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar DH, Ciclitira PJ. Clinical features and diagnosis of celiac disease. Gastroenterology. 2005;128:S19–S24. doi: 10.1053/j.gastro.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Duerkop BA, Vaishnava S, Hooper LV. Immune Responses to the Microbiota at the Intestinal Mucosal Surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DF, Pye G, Bramley R, Clark AG, Dyson TJ, Hardcastle JD. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut. 1988;29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagundes Neto U, Martins MC, Lima FL, Patricio FR, Toledo MR. Asymptomatic environmental enteropathy among slum-dwelling infants. J. Am. Coll. Nutr. 1994;13:51–56. doi: 10.1080/07315724.1994.10718371. [DOI] [PubMed] [Google Scholar]
- Fallingborg J, Christensen LA, Ingeman-Nielsen M, Jacobsen BA, Abildgaard K, Rasmussen HH. pH-profile and regional transit times of the normal gut measured by a radiotelemetry device. Aliment. Pharmacol. Ther. 1989;3:605–613. doi: 10.1111/j.1365-2036.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- Farkas S, Hornung M, Sattler C, Guba M, Steinbauer M, Anthuber M, Herfarth H, Schlitt HJ, Geissler EK. Rapamycin decreases leukocyte migration in vivo and effectively reduces experimentally induced chronic colitis. Int. J. Color. Dis. 2006;21:747–753. doi: 10.1007/s00384-005-0793-7. [DOI] [PubMed] [Google Scholar]
- Fayyaz M, Lackner JM. Serotonin receptor modulators in the treatment of irritable bowel syndrome. Ther. Clin. Risk Manag. 2008;4:41–48. doi: 10.2147/tcrm.s140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenzæ. Br. J. Exp. Pathol. 1929;10:226. [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Fyderek K, Strus M, Kowalska-Duplaga K, Gosiewski T, Wedrychowicz A, Jedynak-Wasowicz U, Sładek M, Pieczarkowski S, Adamski P, Kochan P, et al. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J. Gastroenterol. 2009;15:5287–5294. doi: 10.3748/wjg.15.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RF, Austen WG, Zhang X, Munene G, Mostafa G, Biswas S, McCormack M, Eberlin KR, Nguyen JT, Tatlidede HS, et al. Intestinal alkaline phosphatase is a gut mucosal defense factor maintained by enteral nutrition. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3551–3556. doi: 10.1073/pnas.0712140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al. Human Genetics Shape the Gut Microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345:1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman Y, Tickle TL, Dexheimer PJ, Kim M-O, Tang D, Karns R, Baldassano RN, Noe JD, Rosh J, Markowitz J, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus 2 Emily P, Turnbaugh PJ. Predicting and Manipulating Cardiac Drug Inactivation by the Human Gut Bacterium Eggerthellalenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Harig JM, Soergel KH, Komorowski RA, Wood CM. Treatment of diversion colitis with short-chain-fatty acid irrigation. N. Engl. J. Med. 1989;320:23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, et al. {ACE2} links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Shankar RA, Chzhan M, Samouilov A, Kuppusamy P, Zweier JL. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. 1999;96:4586–4591. doi: 10.1073/pnas.96.8.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity. 2002;17:629–638. doi: 10.1016/s1074-7613(02)00453-3. [DOI] [PubMed] [Google Scholar]
- Henthorn PS, Raducha M, Edwards YH, Weiss MJ, Slaughter C, Lafferty MA, Harris H. Nucleotide and amino acid sequences of human intestinal alkaline phosphatase: close homology to placental alkaline phosphatase. Proc. Natl. Acad. Sci. U. S. A. 1987;84:1234–1238. doi: 10.1073/pnas.84.5.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyworth B, Brown J. Jejunal microflora in malnourished Gambian children. Arch. Dis. Child. 1975;50:27–33. doi: 10.1136/adc.50.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Ianiro G, Bibbò S, Scaldaferri F, Gasbarrini A, Cammarota G. Fecal microbiota transplantation in inflammatory bowel disease: beyond the excitement. Medicine (Baltimore) 2014;93:e97. doi: 10.1097/MD.0000000000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrazábal T, Belcheva A, Girardin SE, Martin A, Philpott DJ. The multifaceted role of the intestinal microbiota in colon cancer. Mol. Cell. 2014;54:309–320. doi: 10.1016/j.molcel.2014.03.039. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific Microbiota Direct the Differentiation of IL-17-Producing T-Helper Cells in the Mucosa of the Small Intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabouridis PS, Lasrado R, McCallum S, Chng SH, Snippert HJ, Clevers H, Pettersson S, Pachnis V. Microbiota Controls the Homeostasis of Glial Cells in the Gut Lamina Propria. Neuron. 2015;85:289–295. doi: 10.1016/j.neuron.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiya M, Silva MJB, Sato K, Ouhara K, Kawai T. Hydrogen mediates suppression of colon inflammation induced by dextran sodium sulfate. Biochem. Biophys. Res. Commun. 2009;386:11–15. doi: 10.1016/j.bbrc.2009.05.117. [DOI] [PubMed] [Google Scholar]
- Kanther M, Tomkovich S, Xiaolun S, Grosser MR, Koo J, Flynn EJ, Jobin C, Rawls JF. Commensal microbiota stimulate systemic neutrophil migration through induction of serum amyloid A. Cell. Microbiol. 2014;16:1053–1067. doi: 10.1111/cmi.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona P, Katona-Apte J. The interaction between nutrition and infection. Clin. Infect. Dis. 2008;46:1582–1588. doi: 10.1086/587658. [DOI] [PubMed] [Google Scholar]
- Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu T-C, Stappenbeck TS, Maleta KM, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Transl. Med. 2015;7:276ra24. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellow JE, Borody TJ, Phillips SF, Tucker RL, Haddad AC. Human interdigestive motility: variations in patterns from esophagus to colon. Gastroenterology. 1986;91:386–395. doi: 10.1016/0016-5085(86)90573-1. [DOI] [PubMed] [Google Scholar]
- Kelly P, Menzies I, Crane R, Zulu I, Nickols C, Feakins R, Mwansa J, Mudenda Vi, Katubulushi M, Greenwald S, et al. Responses of small intestinal architecture and function over time to environmental factors in a tropical population. Am J Trop Med Hyg. 2004;70:412–419. [PubMed] [Google Scholar]
- Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keusch GT, Denno DM, Black RE, Duggan C, Guerrant RL, Lavery JV, Nataro JP, Rosenberg IH, Ryan ET, Tarr PI, et al. Environmental enteric dysfunction: pathogenesis, diagnosis, and clinical consequences. Clin. Infect. Dis. 2014;4(59 Suppl):S207–S212. doi: 10.1093/cid/ciu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khin-Maung-U Bolin TD, Duncombe VM, Myo-Khin Nyunt-Nyunt-Wai Pereira SP, Linklater JM. Epidemiology of small bowel bacterial overgrowth and rice carbohydrate malabsorption in Burmese (Myanmar) village children. Am. J. Trop. Med. Hyg. 1992;47:298–304. doi: 10.4269/ajtmh.1992.47.298. [DOI] [PubMed] [Google Scholar]
- Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpe PS, Petri WA. Environmental enteropathy: critical implications of a poorly understood condition. Trends Mol. Med. 2012;18:328–336. doi: 10.1016/j.molmed.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Meyerson M, Garrett WS. Microbes and inflammation in colorectal cancer. Cancer Immunol. Res. 2013a;1:150–157. doi: 10.1158/2326-6066.CIR-13-0101. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe. 2013b;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen A-M, Peet A, Tillmann V, Pöhö P, Mattila I, et al. The Dynamics of the Human Infant Gut Microbiome in Development and in Progression toward Type 1 Diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantis A. GABA in the Mammalian Enteric Nervous System. News Physiol Sci. 2000;15:284–290. doi: 10.1152/physiologyonline.2000.15.6.284. [DOI] [PubMed] [Google Scholar]
- Larsson JMH, Karlsson H, Crespo JG, Johansson MEV, Eklund L, Sjövall H, Hansson GC. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm. Bowel Dis. 2011;17:2299–2307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- Lecuit M, Abachin E, Martin A, Poyart C, Pochart P, Suarez F, Bengoufa D, Feuillard J, Lavergne A, Gordon JI, et al. Immunoproliferative small intestinal disease associated with Campylobacter jejuni. N. Engl. J. Med. 2004;350:239–248. doi: 10.1056/NEJMoa031887. [DOI] [PubMed] [Google Scholar]
- Lin A, Arnold BF, Afreen S, Goto R, Huda TMN, Haque R, Raqib R, Unicomb L, Ahmed T, Colford JM, et al. Household environmental conditions are associated with enteropathy and impaired growth in rural Bangladesh. Am. J. Trop. Med. Hyg. 2013;89:130–137. doi: 10.4269/ajtmh.12-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens JR, Gurung P, Vogel P, Johnson GR, Carter RA, McGoldrick DJ, Bandi SR, Calabrese CR, Walle L, Vande Lamkanfi M, et al. Dietary modulation of the microbiome affects autoinflammatory disease. Nature. 2014;516:246–249. doi: 10.1038/nature13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. doi: 10.1038/nature13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, Macfarlane S. Bacteria, Colonic Fermentation, and Gastrointestinal Health. J. AOAC Int. 2012;95:50–60. doi: 10.5740/jaoacint.sge_macfarlane. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Marchesi JR, Dutilh BE, Hall N, Peters WHM, Roelofs R, Boleij A, Tjalsma H. Towards the Human Colorectal Cancer Microbiome. PLoS One. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis KG, Stevanovic K, Li Z, Yang QM, Oravecz T, Zambrowicz B, Jhaver KG, Diacou A, Gershon MD. Pharmacological reduction of mucosal but not neuronal serotonin opposes inflammation in mouse intestine. Gut. 2014;63:928–937. doi: 10.1136/gutjnl-2013-304901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle JGM, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McCoy AN, Araujo-Perez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium Is Associated with Colorectal Adenomas. PLoS One. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Wu GD, Lewis JD, Bushman FD. Conservation of gene cassettes among diverse viruses of the human gut. PLoS One. 2012;7:e42342. doi: 10.1371/journal.pone.0042342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár K, Vannay A, Szebeni B, Bánki NF, Sziksz E, Cseh A, Győrffy H, Lakatos PL, Papp M, Arató A, et al. Intestinal alkaline phosphatase in the colonic mucosa of children with inflammatory bowel disease. World J. Gastroenterol. 2012;18:3254–3259. doi: 10.3748/wjg.v18.i25.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal D, Minak J, Alam M, Liu Y, Dai J, Korpe P, Liu L, Haque R, Petri WA. Contribution of enteric infection, altered intestinal barrier function, and maternal malnutrition to infant malnutrition in Bangladesh. Clin. Infect. Dis. 2012;54:185–192. doi: 10.1093/cid/cir807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon C, Baldridge MT, Wallace MA, Burnham C-AD, Virgin HW, Stappenbeck TS. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature. 2015 doi: 10.1038/nature14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-Dependent Crosstalk Between Macrophages and ILC3 Promotes Intestinal Homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Koscsó B, Rajani GM, Stevanovic K, Berres M-L, Hashimoto D, Mortha A, Leboeuf M, Li X-M, Mucida D, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami I, Wakasa Y, Yamashita S, Kurihara T, Zama K, Kobayashi N, Mizutani Y, Mitsutake S, Shigyo T, Igarashi Y. Phytoceramide and sphingoid bases derived from brewer’s yeast Saccharomyces pastorianus activate peroxisome proliferator-activated receptors. Lipids Health Dis. 2011;10:150. doi: 10.1186/1476-511X-10-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y-D, Chang H-W, Kim K-H, Roh SW, Kim M-S, Jung M-J, Lee S-W, Kim J-Y, Yoon J-H, Bae J-W. Bacterial, archaeal, and eukaryal diversity in the intestines of Korean people. J. Microbiol. 2008;46:491–501. doi: 10.1007/s12275-008-0199-7. [DOI] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngure FM, Reid BM, Humphrey JH, Mbuya MN, Pelto G, Stoltzfus RJ. Water, sanitation, and hygiene (WASH), environmental enteropathy, nutrition, and early child development: making the links. Ann. N. Y. Acad. Sci. 2014;1308:118–128. doi: 10.1111/nyas.12330. [DOI] [PubMed] [Google Scholar]
- Van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JFWM, Tijssen JGP, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, et al. Disease-Specific Alterations in the Enteric Virome in Inflammatory Bowel Disease. Cell. 2015;160 doi: 10.1016/j.cell.2015.01.002. http://dx.doi.org/10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Ohama T, Hori M, Fujisawa M, Kiyosue M, Hashimoto M, Ikenoue Y, Jinno Y, Miwa H, Matsumoto T, Murata T, et al. Downregulation of CPI-17 contributes to dysfunctional motility in chronic intestinal inflammation model mice and ulcerative colitis patients. J. Gastroenterol. 2008;43:858–865. doi: 10.1007/s00535-008-2241-2. [DOI] [PubMed] [Google Scholar]
- Olsen I, Jantzen E. Sphingolipids in Bacteria and Fungi. Anaerobe. 2001;7:103–112. [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoike IU, Abiodun PO. Upper small intestinal microflora in diarrhea and malnutrition in Nigerian children. J. Pediatr. Gastroenterol. Nutr. 1989;9:314–321. doi: 10.1097/00005176-198910000-00009. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. Immunoglobulin A Coating Identifies Colitogenic Bacteria in Inflammatory Bowel Disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma GDe, Capilla A, Nova E, Castillejo G, Varea V, Pozo T, Garrote JA, Polanco I, López A, Ribes-Koninckx C, et al. Influence of milk-feeding type and genetic risk of developing coeliac disease on intestinal microbiota of infants: the PROFICEL study. PLoS One. 2012;7:e30791. doi: 10.1371/journal.pone.0030791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Walters WA, Lauber CL, Clemente JC, Berg-Lyons D, Teiling C, Kodira C, Mohiuddin M, Brunelle J, Driscoll M, et al. Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front. Microbiol. 2014;5:298. doi: 10.3389/fmicb.2014.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, Guan J-L, Saitoh T, Akira S, Seglen PO, et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 2013;32:3130–3144. doi: 10.1038/emboj.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen AM, Stensvold CR, Mirsepasi H, Engberg J, Friis-Møller A, Porsbo LJ, Hammerum AM, Nordgaard-Lassen I, Nielsen HV, Krogfelt KA. Active ulcerative colitis associated with low prevalence of Blastocystis and Dientamoeba fragilis infection. Scand. J. Gastroenterol. 2013;48:638–639. doi: 10.3109/00365521.2013.780094. [DOI] [PubMed] [Google Scholar]
- Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: “RePOOPulating” the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch W, Panés J, Lémann M, Schreiber S, Feagan B, Schmidt S, Sturniolo GC, Mikhailova T, Alexeeva O, Sanna L, et al. A multicenter, randomized, double-blind trial of everolimus versus azathioprine and placebo to maintain steroid-induced remission in patients with moderate-to-severe active Crohn’s disease. Am. J. Gastroenterol. 2008;103:2284–2292. doi: 10.1111/j.1572-0241.2008.02024.x. [DOI] [PubMed] [Google Scholar]
- Dos Reis JC, de Morais MB, Oliva CAG, Fagundes-Neto U. Breath hydrogen test in the diagnosis of environmental enteropathy in children living in an urban slum. Dig. Dis. Sci. 2007;52:1253–1258. doi: 10.1007/s10620-006-9288-9. [DOI] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigottier-Gois L. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J. 2013;7:1256–1261. doi: 10.1038/ismej.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/beta-Catenin Signaling via its FadA Adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said HM. Intestinal absorption of water-soluble vitamins in health and disease. Biochem. J. 2011;437:357–372. doi: 10.1042/BJ20110326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge TC, Newman P, Pothoulakis C, Ruhl A, Neunlist M, Bourreille A, Hurst R, Sofroniew MV. Enteric glia regulate intestinal barrier function and inflammation via release of S-nitrosoglutathione. Gastroenterology. 2007;132:1344–1358. doi: 10.1053/j.gastro.2007.01.051. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Stensvold CR, Rajilić-Stojanović M, Heilig HGHJ, De Vos WM, O’Toole PW, Cotter PD. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 2014;90:326–330. doi: 10.1111/1574-6941.12396. [DOI] [PubMed] [Google Scholar]
- Schaible UE, Kaufmann SHE. Malnutrition and infection: complex mechanisms and global impacts. PLoS Med. 2007;4:e115. doi: 10.1371/journal.pmed.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheppach W. Effects of short chain fatty acids on gut morphology and function. Gut. 1994;35:S35–S38. doi: 10.1136/gut.35.1_suppl.s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013:2. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, Sansonetti PJ. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015 doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütte A, Ermund A, Becker-Pauly C, Johansson MEV, Rodriguez-Pineiro AM, Bäckhed F, Müller S, Lottaz D, Bond JS, Hansson GC. Microbial-induced meprin β cleavage in MUC2 mucin and a functional CFTR channel are required to release anchored small intestinal mucus. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12396–12401. doi: 10.1073/pnas.1407597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scupham AJ, Presley LL, Wei B, Bent E, Griffith N, McPherson M, Zhu F, Oluwadara O, Rao N, Braun J, et al. Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 2006;72:793–801. doi: 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL, Garrett WS. Microbes, Microbiota, and Colon Cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, He B, Cassis L, Bigas A, Cols M, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiga H, Kajiura T, Shinozaki J, Takagi S, Kinouchi Y, Takahashi S, Negoro K, Endo K, Kakuta Y, Suzuki M, et al. Changes of faecal microbiota in patients with Crohn’s disease treated with an elemental diet and total parenteral nutrition. Dig. Liver Dis. 2012;44:736–742. doi: 10.1016/j.dld.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013a;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science. 2013b;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- Suply E, de Vries P, Soret R, Cossais F, Neunlist M. Butyrate enemas enhance both cholinergic and nitrergic phenotype of myenteric neurons and neuromuscular transmission in newborn rat colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G1373–G1380. doi: 10.1152/ajpgi.00338.2011. [DOI] [PubMed] [Google Scholar]
- Tan KSW. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin. Microbiol. Rev. 2008;21:639–665. doi: 10.1128/CMR.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkeltaub JA, McCarty TR, Hotez PJ. The intestinal protozoa: emerging impact on global health and development. Curr. Opin. Gastroenterol. 2015;31:38–44. doi: 10.1097/MOG.0000000000000135. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz AJ, Manary MJ, Stephenson K, Agapova S, Manary FG, Thakwalakwa C, Shulman RJ, Manary MJ. Abnormal gut integrity is associated with reduced linear growth in rural Malawian children. J. Pediatr. Gastroenterol. Nutr. 2012;55:747–750. doi: 10.1097/MPG.0b013e3182650a4d. [DOI] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M, Sonnenburg JL, Comstock LE, Bluestone JA, Fischbach MA. Production of α-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, Zimmermann M, Taketani M, Ishihara A, Kashyap PC, Fraser JS, Fischbach MA. Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine. Cell Host Microbe. 2014;16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang J-P, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender Bias in Autoimmunity Is Influenced by Microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CCGM, Troost FJ, Bork P, Wels M, de Vos WM, Kleerebezem M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]