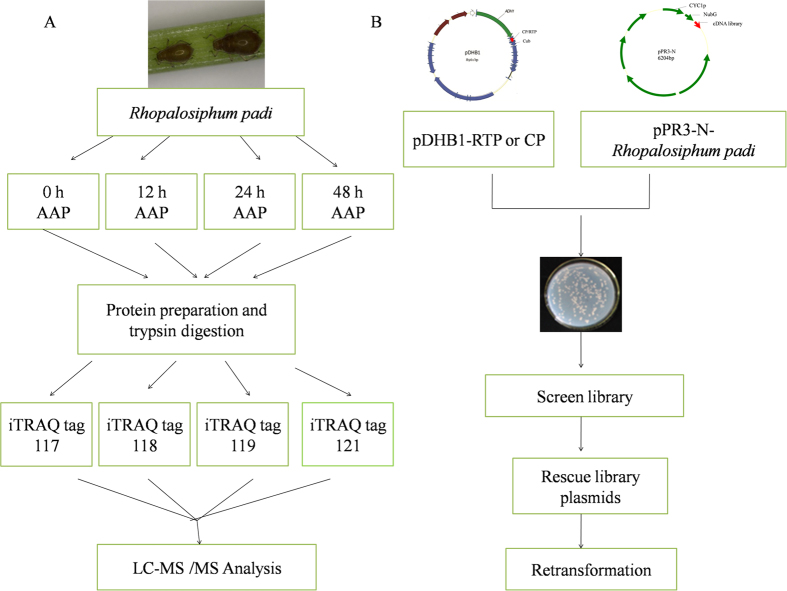
Figure 1. Schematic of iTRAQ design.
(A) and yeast two-hybrid screen design (B). iTRAQ experiment was carried out using the vector aphids Rhopalosiphum padi, which were allowed a 0, 12, 24 or 48 h acquisition access period (AAP) on BYDV-GPV-infected oat plants. Proteins were digested by trypsin, and four iTRAQ labels were used: tag 117 for the 0 h AAP, tag 118 for the 12 h AAP, tag 119 for the 24 h AAP and tag 121 for the 48 h AAP. After labeling, peptides from all four samples were combined and fractionated by chromatography. Each fraction was then analyzed by LC-MS/MS. For yeast two-hybrid screen, the ORFs of BYDV-RTD/BYDV-CP were constructed individually in the yeast expression vector pDHB1, and the cDNA library of R. padi was constructed in prey plasmid pPR3-N. The cDNA library of R. padi was then screened and positive plasmids were rescued. The prey plasmid and bait plasmid were then used to cotransform yeast strain NMY51 to eliminate false positive hits.