Figure 5.
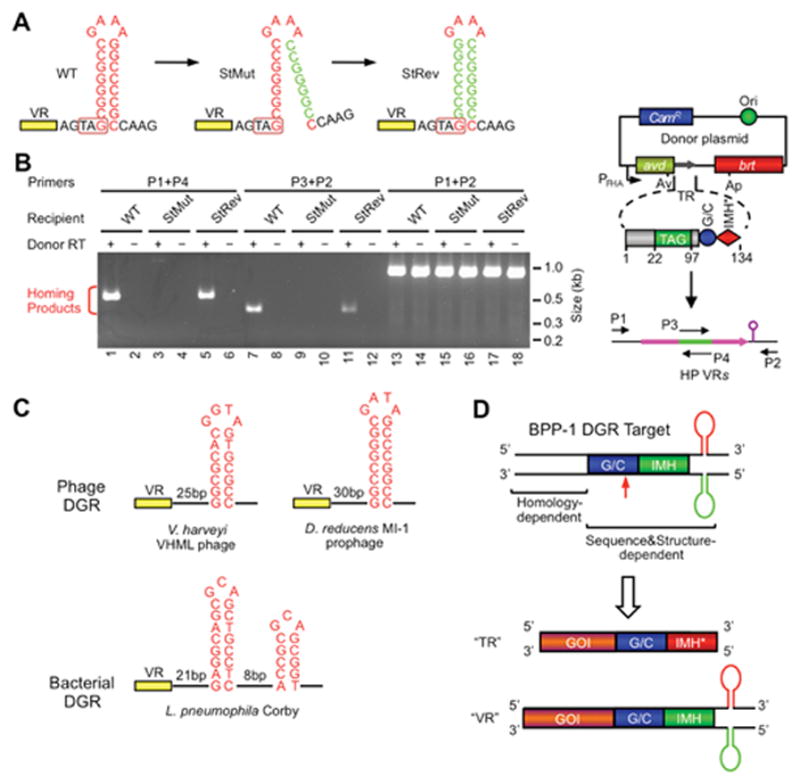
Role of a DNA secondary structure in DGR target recognition. (A) A DNA hairpin/cruciform structure downstream of VR is required for BPP-1 DGR target recognition. The wild type (WT) structure contains an 8 bp GC-rich stem and a 4 nt GAAA loop and is located 4 bp downstream of VR. Mutating the 3′ half of the stem (StMut) dramatically reduced DGR mutagenic homing (B) and phage tropism switching (not shown), while complementary changes to the 5′ half of the stem (StRev) restored DGR activity in both assays. (B) PCR-based DGR homing assays with sequence-tagged TRs and VRs flanked by WT or mutant stem sequences. Shown on the right is a diagram of the PCR assay. Green represents the tag sequence transferred from TR to VR. P1-4 are primers annealing to the tag or flanking regions. (C) Similar DNA structures are found at analogous positions in a number of other phage (two shown) and bacterial (one shown) DGRs. The phage stems are GC-rich and range from 7 to 10 bp, and loops have a conserved 4 nt sequence, G(A/G)NA. The L. pneumophila Corby DGR has a more complex tandem structure that is required for homing. (D) BPP-1 DGR target recognition at the 3′ end is both sequence and structure-dependent, requiring GC, IMH and a hairpin/cruciform structure. Target recognition at the 5′ end is homology-mediated. By inserting a gene of interest (GOI) upstream of GC, IMH and the DNA structure, the heterologous gene can be diversified by the BPP-1 DGR through appropriate engineering of TR. (Adapted from reference 41)
