Figure 6.
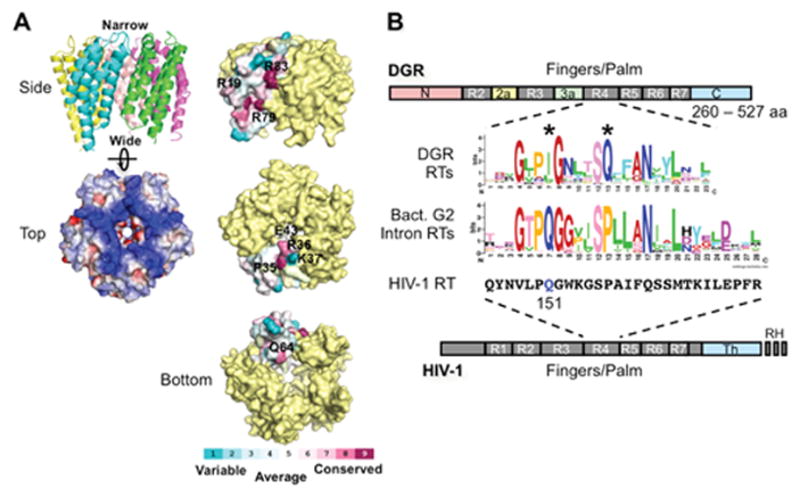
Avd and Brt. (A) (Left) The BPP-1 Avd protein forms a homopentameric structure, with each monomer containing 4 helices running up and down (side view). The pentamer is highly positively charged (top view; blue, positively charged; red, negatively charged). (Right) Amino acid residues on the side, top, and bottom of the Avd pentamer that were tested for Avd-Brt binding and/or DGR homing (27). (B) DGR RT domains and the sequence logo of its highly conserved domain R4. R1-R7 are conserved sequence blocks found in the finger and palm domains of retroviral RTs, such as HIV-1 RT (bottom). DGR RTs contain sequence insertions between R2 and R3 (R2a), and between R3 and R4 (R3a), as well as divergent N- and C-termini. They do not contain the thumb (Th) and RNase H (RH) domains that are found in HIV-1 RT. The domain R4 sequence logo of 155 DGR RTs was generated by Schillinger et al. using WebLogo (13, 49, 50). Comparison with the domain R4 sequence logo that we generated from 93 bacterial group II intron RTs [group II intron database: http://webapps2.ucalgary.ca/~groupii/orf/orfalignment.html; (51–53)], which are most closely related to DGR RTs, showed several characteristic differences, including the two highly conserved positions labeled with *. Also included for comparison is the corresponding amino acid sequence block of HIV-1 RT (Strain BRU; accession # K02013). The glutamine residue at position 151, which plays a role in nucleotide and template preference during reverse transcription, is highlighted in blue. (Adapted from reference 13)
