Figure 5.
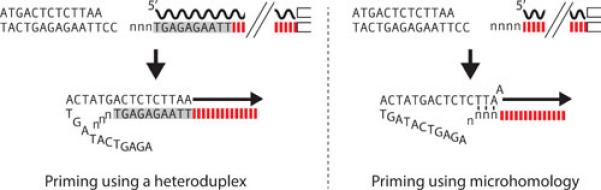
Variation in the priming of second-strand DNA synthesis. R2 elements differ in whether the 5’ end of the RNA template used in the integration reaction ends at the boundary between R2 and the 28S gene or extends a short distance upstream in the 28S rRNA sequence. This difference is dependent upon the location of the self-cleavage site by the R2 ribozyme (see text). Left panel. If self-cleavage by the R2 ribozyme is upstream in the 28S gene sequences, the resulting cDNA strand can form a heteroduplex with the upstream target DNA. This heteroduplex can stabilize the integration intermediate resulting in precise initiation of second strand synthesis (arrow) and uniform 5’ ends for different R2 copies. Right panel. If self-cleavage is at the 28S/R2 junction, there are no 28S sequences on the DNA strand (cDNA) generated by reverse transcription. As a consequence, the R2 protein must use regions of microhomology to initiate second strand synthesis (arrow). Priming frequently involves the 3-5 non-templated nucleotides added to the cDNA strand as the enzyme ran off the RNA template (lower case n's). This use of chance microhomologies to prime second strand DNA synthesis gives rise to sequence variation at the 5’ junctions of different integrated copies of R2. Wavy black line, RNA with 5’ end denoted; red dashed line, first strand DNA composed of R2 sequences; gray box, first strand DNA sequences complementary to upstream DNA target sequences.
