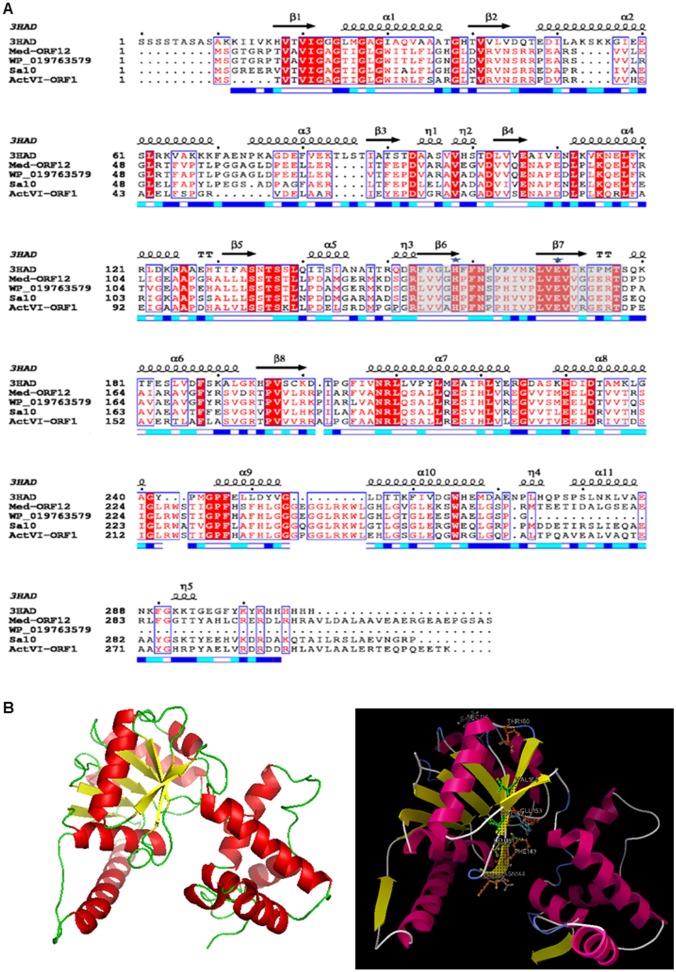
Fig 3. Structure-based sequence alignment between Med-ORF12 and its homologies and the modeled structure of Med-ORF12.
. A: The primary sequence of Med-ORF12 is aligned with the ActVI-ORF1, 3HAD, Sa10 and WP_019763579. The sequences are annotated with corresponding secondary structures in 3HAD. Arrows represent β -sheets and helices indicate α-helices. The similar residues are shaded and identical residues are highlighted in black. The key catalytic residues are marked by ☆. The numbers indicating amino acid positions of each entry refer to the nucleotide sequence deposited in the databases. Sequences retrieved from GenBank were aligned using Clustal X. B: Homology model of Med-ORF12 using one chain of 3HAD from human heart as template. Left: The homology modeled structure of Med-ORF12 obtained using PyMOL. β-strands and α-helices are shown in yellow and red, respectively. Right: The NADH molecule is modeled in active sites of Med-ORF12 by the docking program Autodock. The NADPH is shown in cyans sticks, the catalytic dyad (His141 and Glu153) shown in magenta sticks, the residues (Pro142, Phe143, Asn144, Ser229 and Thr230) interacted by hydrogen bonding with NADH shown as sky-blue sticks and the hydrogen bonds shown in blue dots.