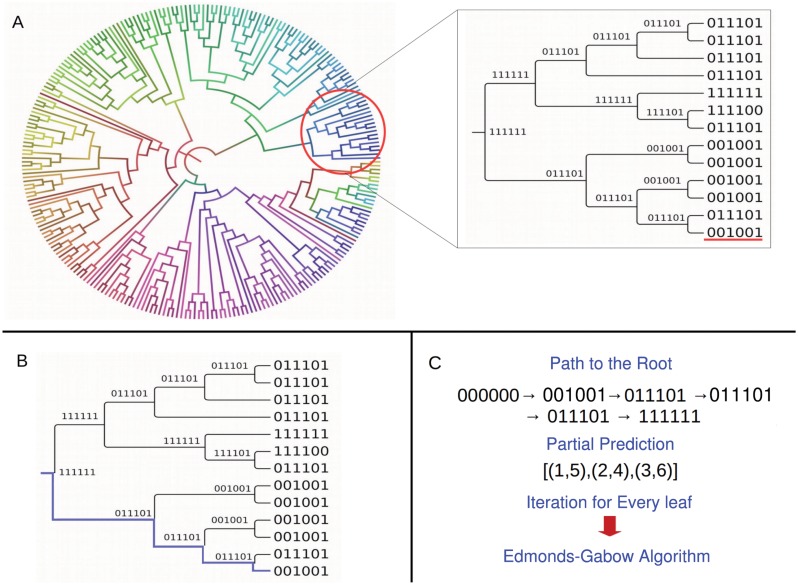
Fig 3. Illustrative example of the PhyloCys algorithm for inferring disulfide bonds from patterns of co-variation among lineages.
Given a phylogenetic tree and the binary labels assigned to each inner node (A), the path from the last leaf to the root is highlighted in (B). The sequence of nodes encountered from the leaf to the root (C) is used to assign weights to the connectivity graph and the most likely connectivity pattern is obtained with Edmonds-Gabow algorithm (C).