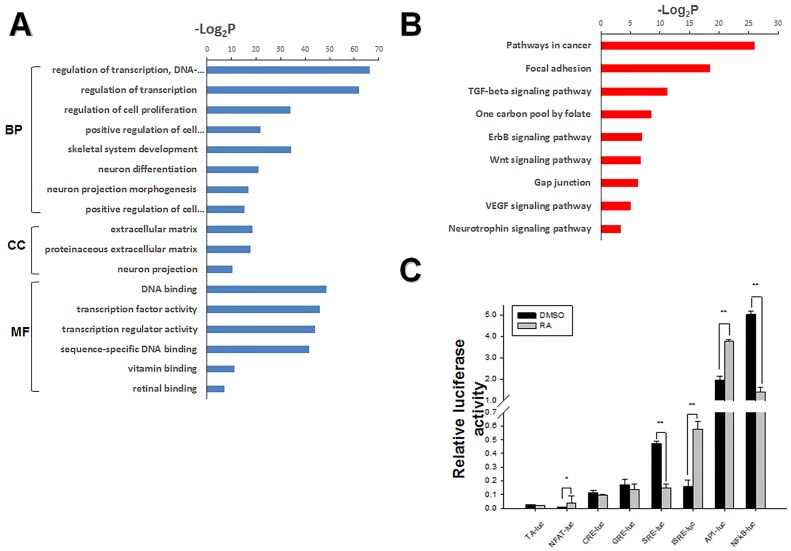
Fig 3. GO annotation and Pathway screening to identify signaling pathways involved in RA-induced differentiation of mESCs.
(A) GO categories for comparison of differently expressed genes identified by microarray analysis. Using the DAVID online system, significantly regulated genes were grouped into three main GO categories: biological processes (BP), cellular components (CC), and molecular functions (MF) (shown as a—log2P value). Significantly regulated genes according to fold change (FC) in gene expression (FC ≥ 2 or FC ≤ 0.5). (B) KEGG pathway categories of significantly differentially expressed genes (2225) in mESCs after RA stimulation. (C) Signaling pathway profiling of mESCs in response to RA stimulation for 24 h. Cells co-transfected with reporter plasmids and internal control pRL-SV40. Dual luciferase assays of pathway reporters showed that NF-kB, NFAT, SRE, and AP1 signaling were significantly activated by RA. Data represented as mean ± SEM of triplicate experiments. Pathway reporters used for screening and their represented pathways. Reporter plasmids for signaling pathways involving PKC and Ca2+/calcineurin (pNFAT-luc), PKA (pCRE-TA-luc), glucocorticoid/HSP90 (pGRE-TA-luc), MAPK/JNK (pSRE-TA-luc), proliferation/inflammation (pISRE-TA-luc), NFκB (pNFκB-TA-luc), and JNK/p38 (pAP1-TA-luc). RA (1 μM) or equal volume DMSO was added to the mESCs for 24 h, followed by cell lysis. Luciferase activity is expressed relative to that of pTA-luc. The values for relative luciferase activities are shown in the bar graphs (*p < 0.05; **p < 0.01).