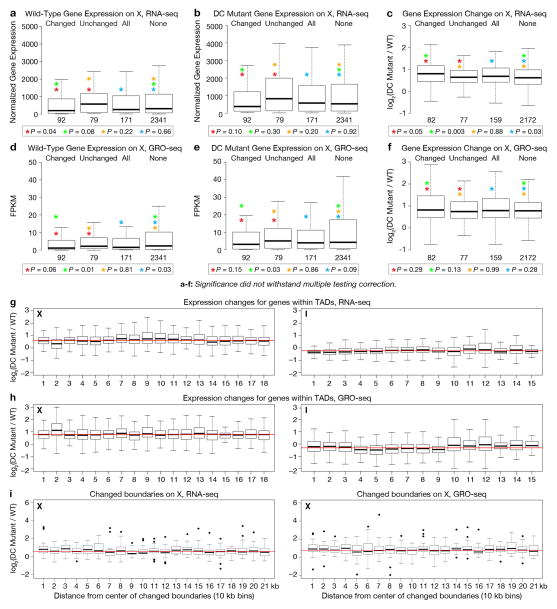
Extended Data Figure 10. DCC-dependent TADs influence global rather than local gene expression.
Gene expression analysis was assayed using RNA-seq or GRO-seq, as indicated. a–b, Boxplots depict expression levels for wild-type or DC mutant embryos assayed by RNA-seq for X genes at changed TAD boundaries, unchanged TAD boundaries, all TAD boundaries or genes not at TAD boundaries. Expression levels are given as normalized read number per kilobase of gene length. c, Boxplots depict the fold change in expression assayed by RNA-seq between wild-type and DC mutant embryos for genes at changed TAD boundaries, unchanged TAD boundaries, all TAD boundaries or genes not at boundaries. The lowest-expressing genes (bottom 10%) were removed from analysis. d–f, As in a–c, but assayed by GRO-seq with gene expression levels given as fragments per kilobase of transcript per million mapped reads (FPKM). For a–f, P-values were calculated using the Mann-Whitney U Test; significance did not withstand multiple testing correction. g–h, Boxplots depict the fold change in the gene expression between wild-type and DC mutant embryos based on RNA-seq or GRO-seq for X and I. Each box has genes from one TAD on X (left) or I (right). Lowest-expressing genes (bottom 10%) were removed from analysis. No discernible pattern was evident for expression changes versus gene location. i, Boxplots depict the fold change in X gene expression between wild-type and DC mutant embryos relative to the distance from the TAD boundary. Each box contains genes in 10 kb bins radiating out from the center of each TAD boundary. The lowest-expressing genes (bottom 10%) were removed from analysis. No discernible pattern to the gene expression changes exists, as assayed by RNA-seq (left) or GRO-seq (right). Weak significance and lack of concordance between RNA-seq and GRO-seq data suggest no biologically relevant correlation between TAD boundaries and local regulation of gene expression.