Fig. 1.
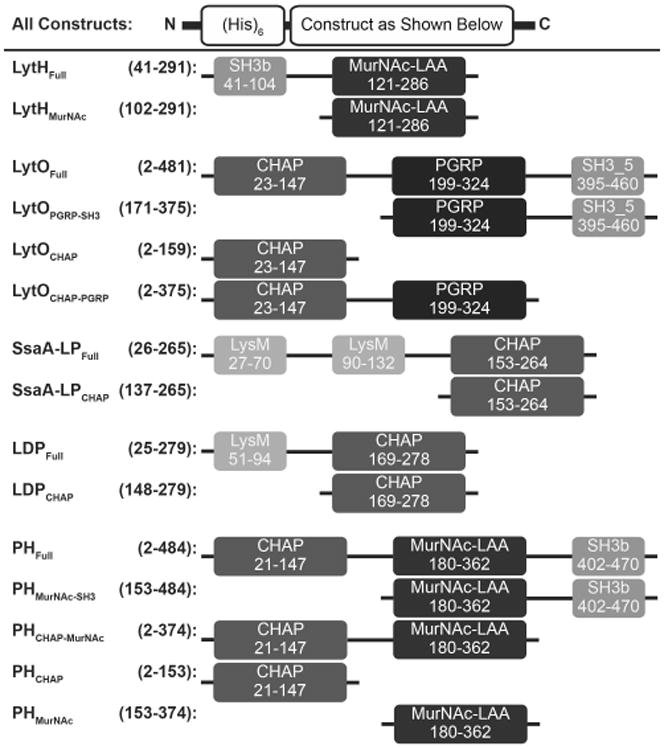
Schematic of autolysin constructs. Numbers represent amino acid positions corresponding to the full length, wild type protein from UniProt. On the left is the construct name, followed by the corresponding amino acid numbers of the truncations. On the right is the illustration of each construct. Domain names and amino acid numbers were obtained from the NCBI Conserved Domain Search and abbreviations are as follows: CHAP, cysteine/histidine-dependent amidohydrolase/peptidase; MurNAc-LAA, N-acetylmuramoyl-L-alanine amidase; PGRP, peptidoglycan recognition proteins; SH3, bacterial SRC homology 3 domain; LysM, lysin motif. Dark greys represent purported catalytic domains and light greys represent purported cell wall binding domains. The function of the PGRP domain, black, is currently unknown.
