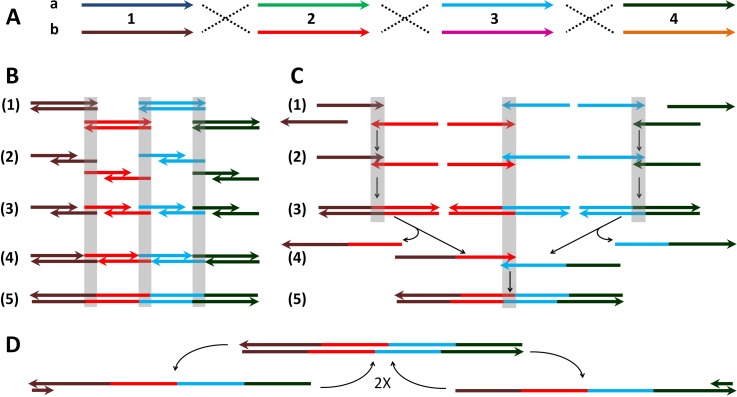
FIG. 4.
DNA library combinatorics, assembly and PCR amplification. The mechanism is illustrated with the example of the library member, 1b2b3a4a, from the combinatorial assembly in (A), which was assembled either isothermally or by thermocycling ((B) and (C)), amplified by PCR (D) and sequenced by cloning.28 (A) Design of combinatorial library. Four pairs ((a) and (b)) of DNA ultramers (150mers: 1, 2, 3, and 4) were designed to make the combinatorial library of 24 = 16 full length DNA sequences (525 nt) via overlap subsequences (length of 25 nt, identical for both members of pair). Sequences were otherwise chosen randomly with minimal complementarity (≤8 successive matches). The details of the sequences employed are shown in supplementary Table 1S.28 Arrows indicate the 5′ to 3′ direction of the DNA. (B) Gibson assembly mechanism. (1) The separate dsDNA inputs are shown aligned via hybridizing overlaps (grey boxes). (2) The exonuclease partially digests the strands from the 3′ ends, by sufficient bases to allow strand hybridization. (3) The newly created single stranded tails containing the 25mer overlapping sequences hybridize to form an assembly complex. (4) DNA polymerase fills the gaps between the pre-assembled strands. The 525 nt complex is formed with 6 nicks (on both strands). Exonuclease becomes inactivated before the end of the reaction and cannot compete further with the polymerase. (5) The nicks are ligated by DNA ligase to complete a full-length dsDNA assembly. (C) Assembly PCR mechanism without added primers: (1) Denaturing of the dsDNA 150 bp inputs (95 °C) or ssDNA input. The strands are shown pre-aligned according to the overlaps as in (B). (2) Assembly of pairs of the ultramers via the 25-mer overlapping sequences (50 °C). (3) Extension of the overlaps by polymerase (72 °C). (4) Following thermal cycle: The denaturing step releases the 1-2 and 3-4 strands containing the 2-3 overlap with subsequent assembly of the strands via annealing (50 °C). Formation of the assemblies 1-2-3 as well as 2-3-4 is also possible, depending on whether the initial inputs 2 and 3 are added in ds or complementary ss form. (5) Extension of the overlaps by polymerase (72 °C). (D) PCR exponential amplification of the assembled 525 bp product using primers 1F (left) and 4F (right). The primed PCR procedure included similar steps as in the assembly PCR (C): denaturing (95 °C), annealing (47 °C), as well as extension (72 °C). See Sec. II B 2 for further details.