Figure 1.
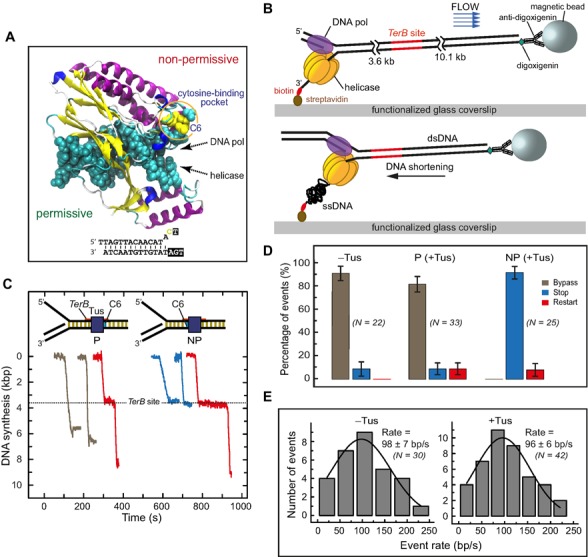
Single-molecule DNA synthesis by T7 helicase–polymerase upon encountering the permissive or non-permissive face of the Tus–TerB complex. (A) Crystal structure (PDB code: 2I06) of the locked Tus–Ter complex shows the flipped C(6) base at the non-permissive face (5). (B) A schematic representation of the single-molecule tethered-bead experimental setup for observing DNA synthesis by the T7 helicase–polymerase. DNA synthesis converts the surface-tethered dsDNA to ssDNA, which at a regime force of 2.6 pN results in shortening of the DNA and displacement of the bead in the opposite direction to the flow. (C) Representative trajectories of DNA synthesis upon encountering Tus–TerB complexes. The fork primarily bypassed Tus–TerB complex on arrival at the permissive face (P, left), while it fully stopped at the non-permissive face (NP, right). Trajectories show permanent stoppage (in blue), unimpeded bypass (gray) or restart (red) at P or NP Tus–TerB. The location of the TerB site is indicated. Traces have been offset on the time axis for clarity. (D) The percentage of populations of replication forks that bypassed, transiently stopped or fully stopped at P TerB and NP TerB are shown. A control experiment in the absence of Tus is shown for NP TerB. Error bars correspond to the standard deviation of binomial distributions. (E) Rate of leading strand synthesis using forked-λ DNA in the absence (left) or presence of Tus (right). The rate distributions were fit (lines in black) with a Gaussian distribution. The uncertainties correspond to the standard error of the distribution. Leading-strand replication reactions were carried out in the presence or in the absence of Tus protein in buffer containing 50 mM potassium glutamate.
