Figure 5.
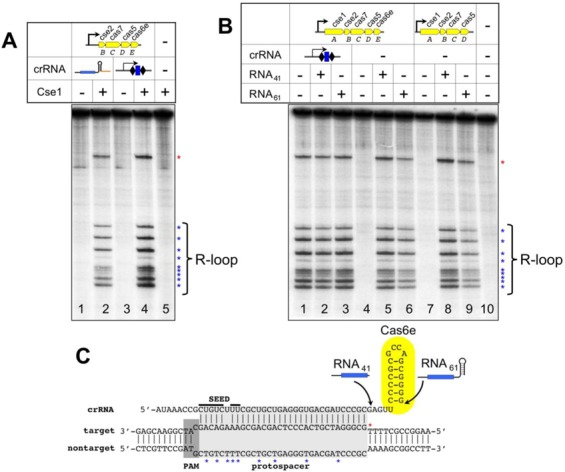
Formation of R-loops in vitro. (A) Cascade was affinity purified from E. coli cells lacking genomic CRISPR arrays, expressing all Cascade subunits but Cse1 and harboring the pG8_crRNA plasmid producing unit-sized crRNA transcript (lanes 1 and 2), or a plasmid containing a CRISPR array with multiply repeated g8 spacer (lanes 3 and 4). Where indicated, Cascade preparations were supplemented with recombinant Cse1, combined with terminally labeled double-stranded DNA fragment containing the g8 protospacer and functional PAM and subjected to potassium permanganate probing. Reaction products were resolved by denaturing urea gel and revealed by phosphoimagery. (B) Cascade was purified from cells lacking any source of crRNA (lanes 4–6) or cells that lacked both the crRNA source and the cas6e gene (lanes 7–9). In lanes 1–3, control Cascade purified from cells harboring a plasmid containing a CRISPR array with multiply repeated g8 spacer was used. Where indicated, Cascade preparations were supplemented with chemically synthesized g8 crRNA (RNA61) or its truncated variant lacking the 3′ handle (RNA41) and used in permanganate probing reactions set up and analyzed as described for panel A. (C) Summary or permanganate probing. The R-loop structure formed on the g8 protospacer target DNA is shown. Asterisks indicate permanganate-sensitive thymine positions in the R-loop complex. The end points of synthetic crRNA molecules used are indicated. The 3′ repeat handle is shown within a yellow oval that represents a tightly bound Cas6e molecule.
