Figure 6.
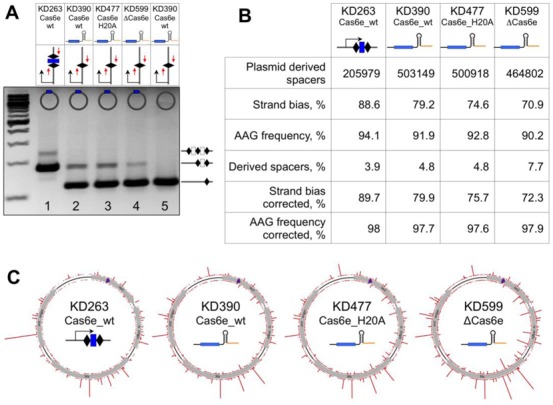
Primed CRISPR adaptation in cells producing unit-sized crRNA transcripts with or without Cas6e. (A) Results of primed adaptation experiment (see ‘Materials and Methods’ section) with indicated cells are shown. The leader-proximal end of CRISPR cassette was amplified using an appropriate primer pair, amplification products were separated by agarose gel electrophoresis and revealed by ethidium bromide staining. (B) DNA fragments corresponding to expanded CRISPR cassettes shown in (A) were subjected to Illumina sequencing. Statistics for reads corresponding to spacers derived from priming protospacer-containing plasmid is presented. ‘Derived spacers’ refers to spacers that originate from recognition of protospacers with correct AAG PAM but are either excised from target DNA with short shifts in the upstream or downstream direction or are inverted during acquisition in CRISPR array. (C) Mapping of spacers acquired in (A) on the priming protospacer plasmid is schematically shown. The position of the priming protospacer (purple triangle) as well as known features of the plasmid are shown. The heights of red bars indicate the efficiency (number of times) a spacer arising from a protospacer in this position was observed. Red bars protruding inside and outside of circles indicating the protospacer plasmid show spacers derived from different strands of DNA.
