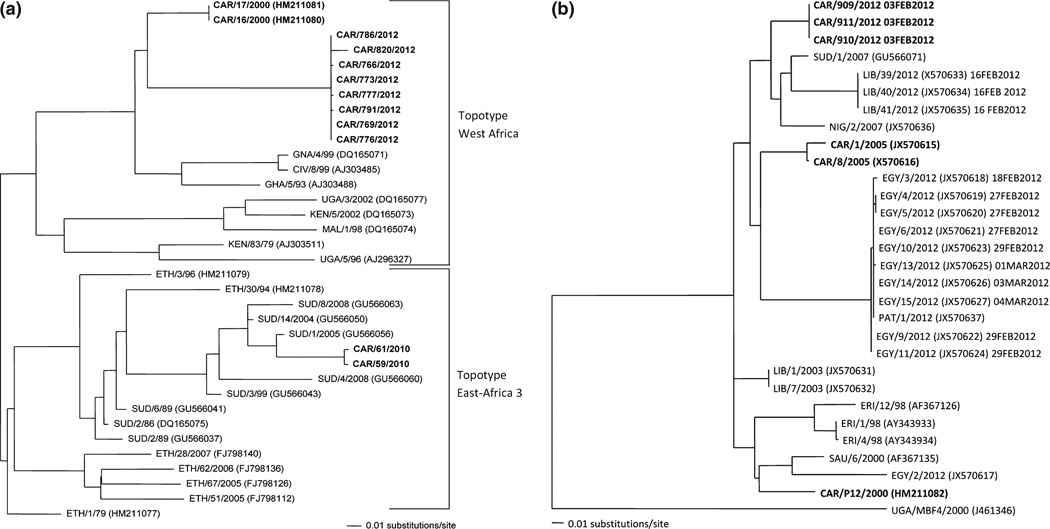
Fig. 5.
Maximum likelihood phylogenetic trees of Cameroon serotype O (a) or SAT2 isolates (b). The phlyogenetic trees are based on 633 nucleotides of VP1. The mid-point tree shown in (a) highlights the serotype O sequences from Cameroon (CAR). West Africa (WA) topotype Cameroon viruses are from 2000 and 2012 and the East Africa (EA) topotype viruses are from 2010. The serotype SAT2 phylogenetic tree (b) above is rooted by UG/V MBF4/2002. Cameroon (CAR) samples are in bold. This phylogenetic tree is of the complete VP1 region. Two recent FMDV outbreaks are of interest: Libya and Egypt shown in blue and green, respectively. The dates of when these samples as well as the SAT2 viruses sequenced for this study have been added. Trees were generated by maximum likelihood algorithm (within PAUP) using the evolutionary model TRN+I+G, no bootstrap values are generated by this method.