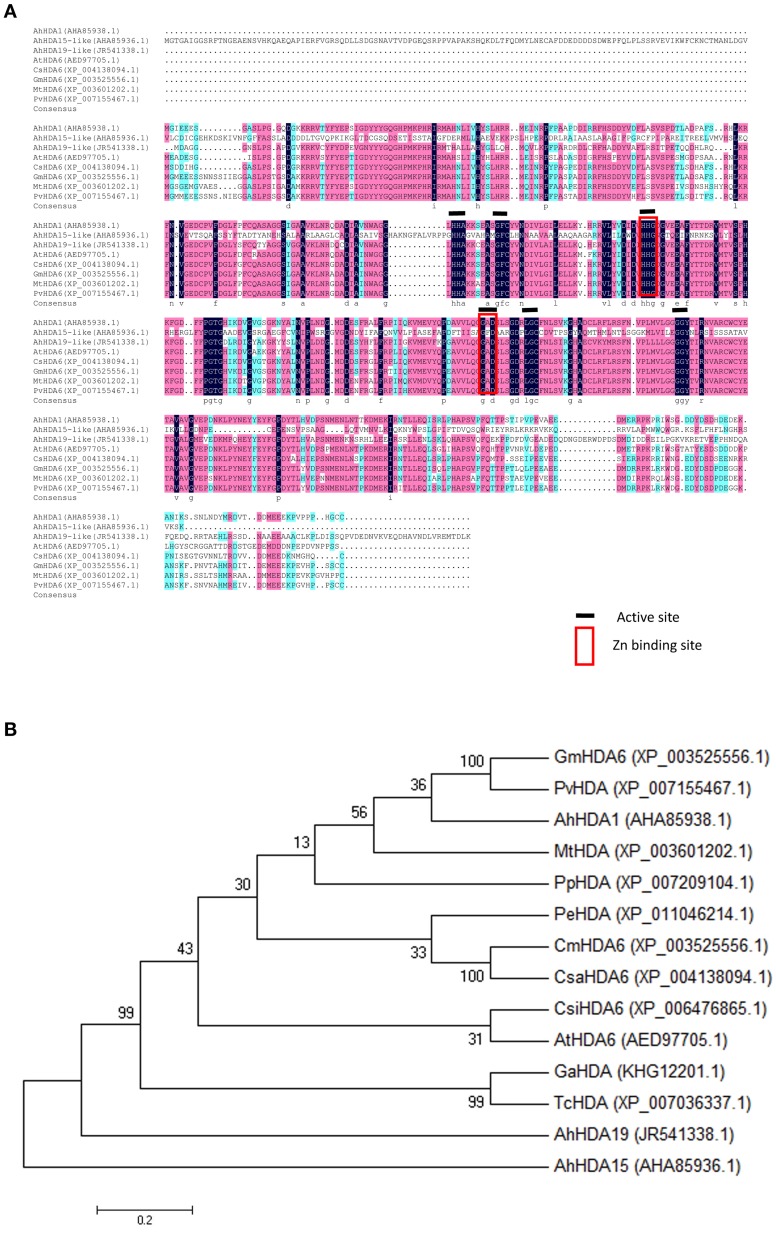
Figure 2.
Relatedness of peanut HDACs sequences to counterparts in other plants. (A) Alignment of deduced amino acid sequence of peanut HDACs with other plant HDAC sequences. The degree of similarity of the amino acid residues at each residues at each aligned position is shaded black, red, blue, in decreasing order. GenBank accession numbers for each aligned HDAC protein are indicated in parenthesis. (B) Phylogenetic analysis of amino acid sequences of AhHDA1 and other plant HDACs. A multiple sequence alignment was performed using Clustal W and the phylogenetic tree was constructed via the Neighbor-Joining method in MEGA 4 software. Bootstrap values from 1000 replicates for each branch are shown. GenBank accession numbers: Glycine max HDA6 (XP_003525556.1), Phaseolus vulgaris HAD (XP_007155467.1), Arachis hypogaea HDA1 (JR541338.1), Medicago truncatula HDA (XP_003601202.1), Prunus persica HDA (XP_007209104.1), Populus euphartica HDA (XP_011046214.1). Cucumis melo HDA6 (XP_00864523.1), Cucumis sativas HDA6 (XP_004138094.1), Citrus sinensis HDA6 (XP_006476865.1), Arabidopsis thaliana HA6 (AED97705.1), Gossypium arboretum HDA (KHG12201.1), Theobroma cacao HDA (XP_007036337.1), Arachis hypogaea HDA19-like (AHA85936.1), Arachis hypogaea HDA15-like (AHA85936.1). The scale bar is 0.02.