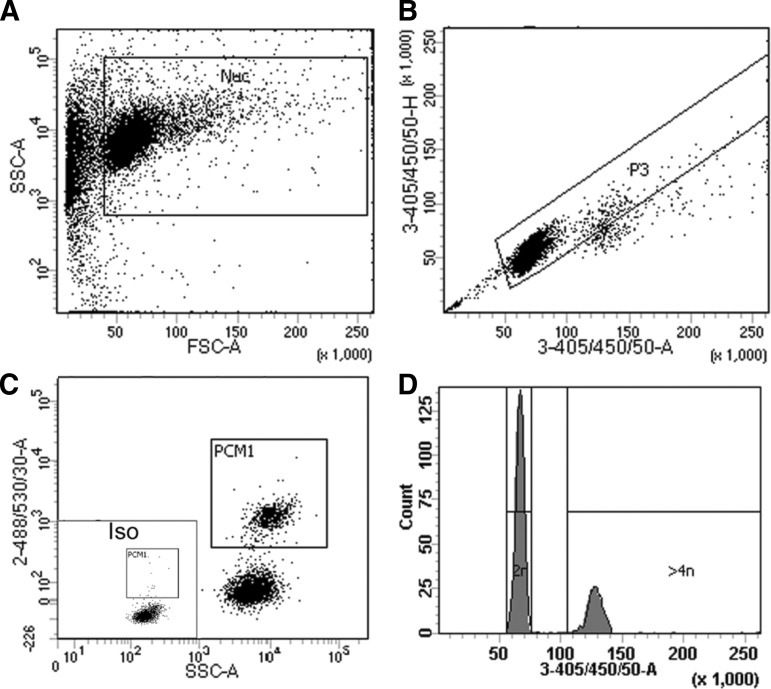
FIG. 2.
Gating strategy for cardiomyocyte nuclei and analysis of ploidy. (A) Cardiomyocyte nuclei are distinguished from debris using a plot of forward scatter versus side scatter. (B) As it is important that only single nuclei are included in subsequent analysis, single nuclei are identified based on 3-355/405/50-area versus 3-355/405/50-height [after 4′,6-diamidino-2-phenylindole (DAPI) labeling]. (C) Isotype control allows gating for the identification of pericentriolar material 1-positive (PCM1+) cardiomyocyte nuclei and PCM1− noncardiomyocyte nuclei. (D) A histogram plot was created using 3-355/405/50-area on a linear scale for the analysis of the ploidy of each individual nucleus. As DAPI binding to DNA is stoichiometric, the fluorescence intensity is proportional to the amount of DNA. The majority of cardiomyocytes are 2N as expected; 4N nuclei have a fluorescent intensity twice that of the 2N population, and 8N nuclei are twice as intense again. All nuclei with an intensity greater than that of the 2N population were excluded from the analysis of cardiomyocyte renewal.