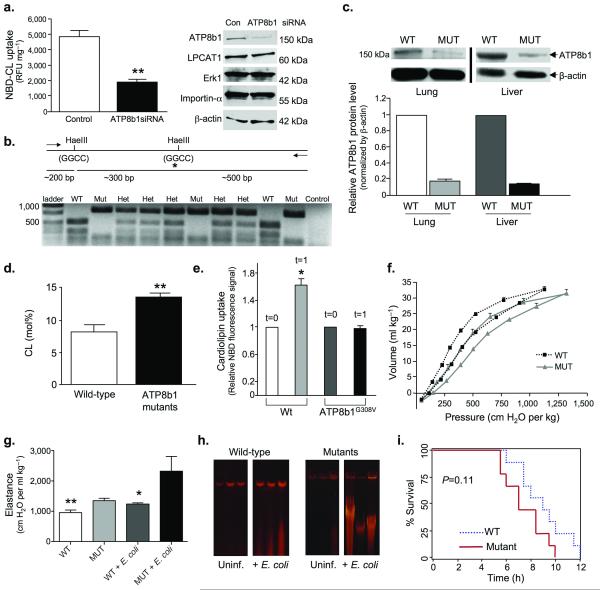
Figure 5. ATP8b1 defective mice are prone to bacterial-induced lung injury.
a. ATP8b1 siRNA knockdown. Human A549 ATII cells were transfected with ATP8b1 siRNA or control RNA and incubated with NBD-labeled CL or PS for 30 min at 37 °C prior to harvest for uptake (left) and immunoblotting (right) using 25 μg of protein loaded/lane. Blots were probed with ATP8b1, LPCAT1, Erk1, importin-α, and β-actin antibodies. **P<0.01. b. ATP8b1G308V/G308V mutants genomic DNA has a Hae III restriction site (GGCC) in which the second G is mutated to T (glycine to valine) and is not recognized in restriction digests. Mutant genomic DNA generates an 800 bp fragment upon HaeIII digestion compared to a 500 bp fragment with wild-type genomic DNA. The HaeIII digestion pattern was used for genotyping wild-type (Wt), heterozygous (Het), and mutant (Mut) mice. c. ATP8b1 immunoblotting in mouse tissues. Below: densitometric analysis from n=six mice/group. d. CL was assayed in lung lavage from ATP8b1 mutant and wild-type littermates (three/group). **P<0.01. e. Primary type II epithelia isolated from mutants or wild-type littermates (five mice/group) were incubated with NBD-CL and cellular uptake was assayed initially after labeling (t=0) and after 1 min. f-g. Mutants and wild-type littermates uninfected or infected (seven/group) with E. coli at 106 CFU/mouse for 48 h were analyzed for lung mechanics. Mice were mechanically ventilated for determination of quasi-static volume-pressure curves (f, [uninfected]), and elastance (g). Statistical significance was determined by a one-way ANOVA where in (g) ** WT vs. MUT + E.coli, P<0.01 and * WT+ E. coli vs. MUT+ E. coli, P<0.05. h. Wild type and mutant mice treated as in (g) were also analyzed for lung DNA fragmentation. i. Kaplan-Meier survival curve for wild-type and ATP8b1 defective mice infected with E. coli, (5 × 106 CFU/mouse, seven mice/group, P=0.11, log rank test).