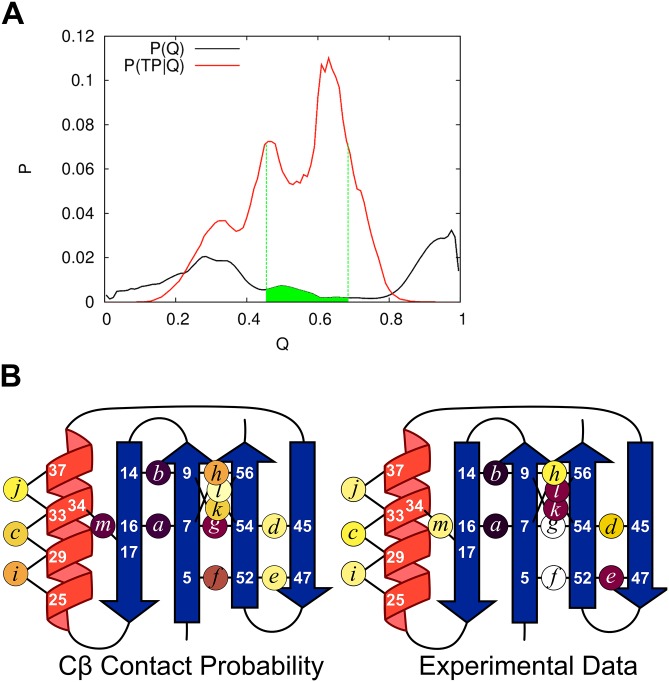
Fig. S7.
Quasi-PFold analysis of the MD trajectories on NuG2b. (A) Selection of the TSEMD begins by calculating P(TP|Q), i.e., the probability of having Q native contacts across the 27 TPs. Candidate TSE structures are selected according to P(TP| 0.45 < Q < 0.68, green). For each such candidate structure, the number of structures is identified from all other structures in the trajectory that have Cα-RMSD < 2 Å but are not in any TP. If at least 10 such structures can be found, PFold is calculating as the fraction of the 10+ structures that folds to the native basin within 1 μs. The TSE is composed of all structures having 0.4 < PFold < 0.6. (B) Cβ contact probability of the TSEMD obtained from Quasi-PFold analysis using the same color scale as Fig. 1. The ensemble displays the N-terminal hairpin as folded with a contacting β4 in its native parallel orientation, and the helix is found to be partially formed. Overall, despite the challenges in reproducing full experimental data even with today’s state-of-the-art all-atom molecular dynamics simulations (partially due to the deficiencies in modern force fields), 19% of the TSEMD structures feature all four strands arranged in a native-like conformation, consistent with the four strands being present in 5 of 27 transition paths.