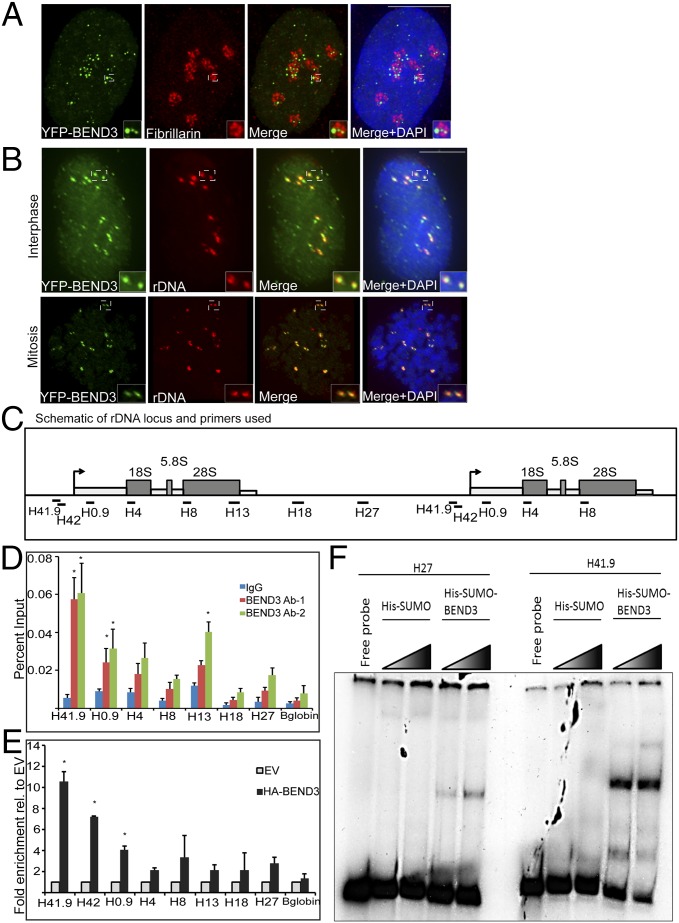
Fig. 1.
BEND3 localizes to rDNA. (A) Immunostaining of the nucleolar marker Fibrillarin (red) in YFP-BEND3 transfected cells. (B) rDNA FISH analysis (red) in YFP-BEND3 transfected U2OS cells during interphase and mitosis. DNA is counterstained with DAPI. (Scale bar: 10 µm.) (C) A schematic of 1.5 units of the human rDNA repeat and primers used for ChIP analysis. H41.9 and H42 are at the promoter; H4, H8, and H13 span the coding region whereas H18 and H27 are at the IGS region. (D) ChIP analysis showing BEND3 occupancy at the rDNA locus. Results are plotted as percentage of input. (E) ChIP analysis showing HA-BEND3 occupancy at the rDNA locus. HA-ChIP was performed in cells stably expressing either an empty vector (EV) or HA-tagged BEND3. Results are plotted as percentage of input values normalized to EV control. Error bars represent SD; n = 3. *P value < 0.05. (F) Mobility shift assay showing His-SUMO-BEND3 binding to rDNA promoter (H41.9) and IGS (H27) sequences in vitro.