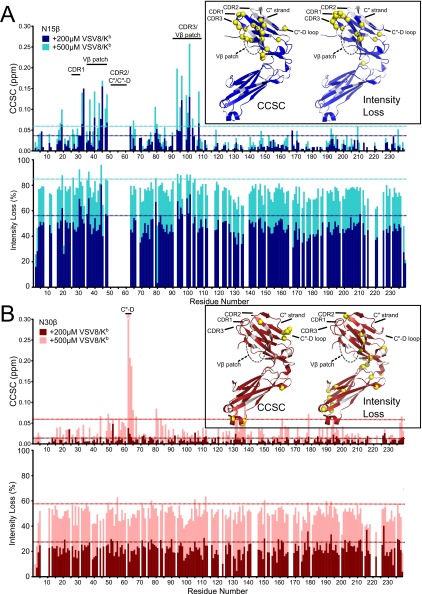
Fig. S2.
Chemical shift perturbation and peak intensity analysis of N15β– and N30β–VSV8/Kb interaction. The 2D 1H-15N HSQC spectra of (A) 15N-N15β or (B) 15N-N30β were acquired alone or with 200 (dark blue or red bars) or 500 μM (light blue or red bars) VSV8/Kb. (Upper) Chemical shift change (CCSC) was calculated for each residue. Dotted lines are color-coded like bars and indicate 1 SD above median CCSC. (Lower) Intensity loss is plotted relative to unligated β. Dotted lines indicate 1 SD above median intensity loss. (A) N15β shows significant changes on addition of VSV8/Kb. Highlighted are changes in CDR1, CDR3, and the Vβ patch regions, which are progressively larger with the addition of more VSV8/Kb. The CDR2, C″ strand, and C″D loop are unassigned. Inset shows ribbon representation of (Left) CCSC analysis or (Right) intensity loss of VSV8/Kb interaction with N15β. Amide protons exhibiting significant (Left) CCSCs or (Right) intensity losses are shown in yellow spheres. Regions corresponding to unassigned residues are white. CCSC data and structural representations in Inset are reproduced from Fig. 2A to aid comparison with N30β. (B) N30β shows nonspecific changes distributed throughout the molecule. Highlighted are C″D loop residues, which show anomalously large CCSCs for the 500 μM level of VSV8/Kb. This anomaly may be caused by a local ring current shift and does not seem to indicate a large binding surface, because very few residues are affected; also, intensity losses are distributed evenly throughout the molecule. *Residue S63, which has a CCSC = 0.44 ppm. The detail of the N30β–VSV8/Kb interaction is the same as in A.