FIGURE 1.
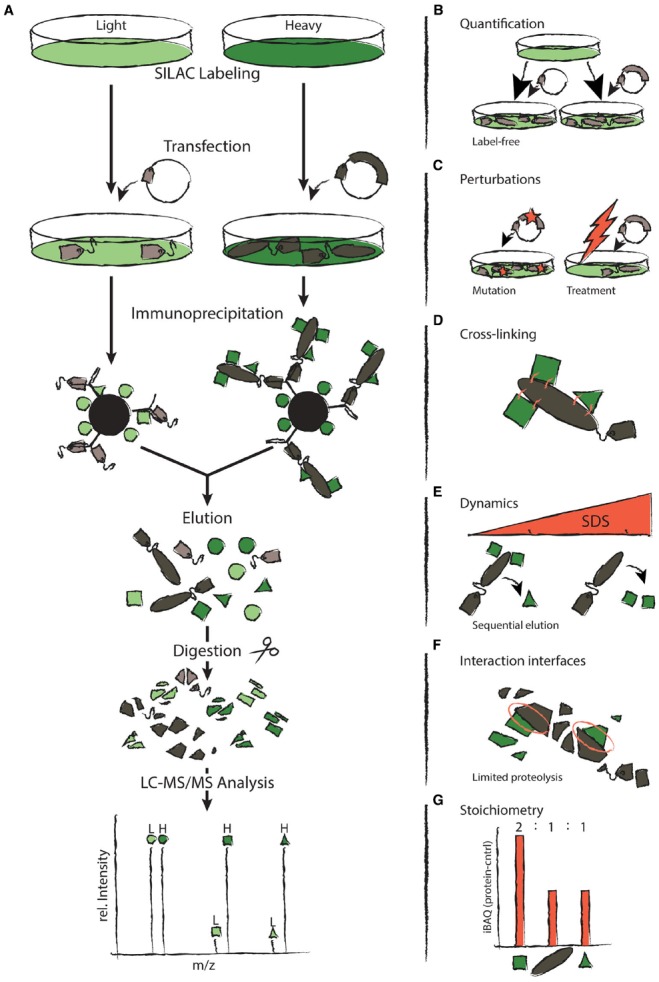
Approaches to q-AP-MS experiments. (A) The left hand side depicts the typical workflow of a SILAC-based q-AP-MS experiment. Differentially SILAC-labeled cells are transfected with a tagged protein of interest or a control vector containing only the tag, respectively. Proteins are immunoprecipitated with antibodies directed against the tag. Samples are mixed prior to elution. Eluted proteins are cleaved into peptides and analyzed by Liquid-Chromatography Mass Spectrometry (LC-MS). (B–G) The right hand side depicts how q-AP-MS can be employed to study different aspects of PPIs. (B) Label-free quantification provides an alternative to SILAC. (C) Immunoprecipitation can compare changes in PPIs upon perturbation. (D) Transient interactions and complex structure can be studied by cross-linking. (E) Submodule composition and PPI dynamics can be revealed by sequential elution with increasing concentrations of SDS. (F) Limited proteolysis provides a means to detect interaction interfaces. (G) The stoichiometry of complexes can be revealed by comparing abundances of the different subunits.
