Figure 2.
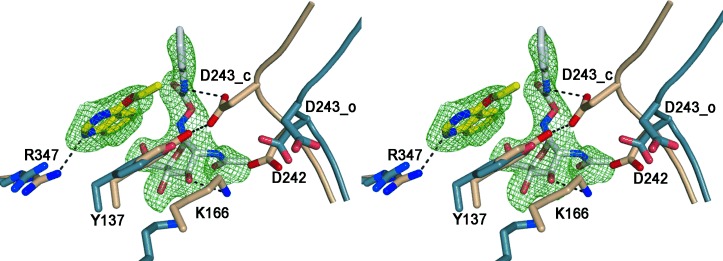
Stereo image of the activator and PUGNAc binding sites on BtGH84. The protein backbone of the catalytic loop is shown schematically with key active site residues in stick representation. The activator 2-bound structure (4UR9) is shown in fawn and the “loop open” structure (formed in the presence of streptozotocin which does not induce the closed conformation) is shown in blue (2W4X), overlaid on common secondary structure. The ligands are shown in stick representation, with carbon atoms of PUGNAc (1) in white and of activator 2 in yellow with the SA-Fo-Fc omit map contoured at 3σ r.m.s. (root mean square). The activator stacks on top of Y137 and forms one H-bond to R347 (H-bonds shown as dashed lines) thereby stabilizing the hydrogen bond between Y137 and D243. In the open structure (blue) the loop containing the catalytic residues D242 and D243 is shifted away from the active site and the H-bond between Y137 and D243 is lost.
