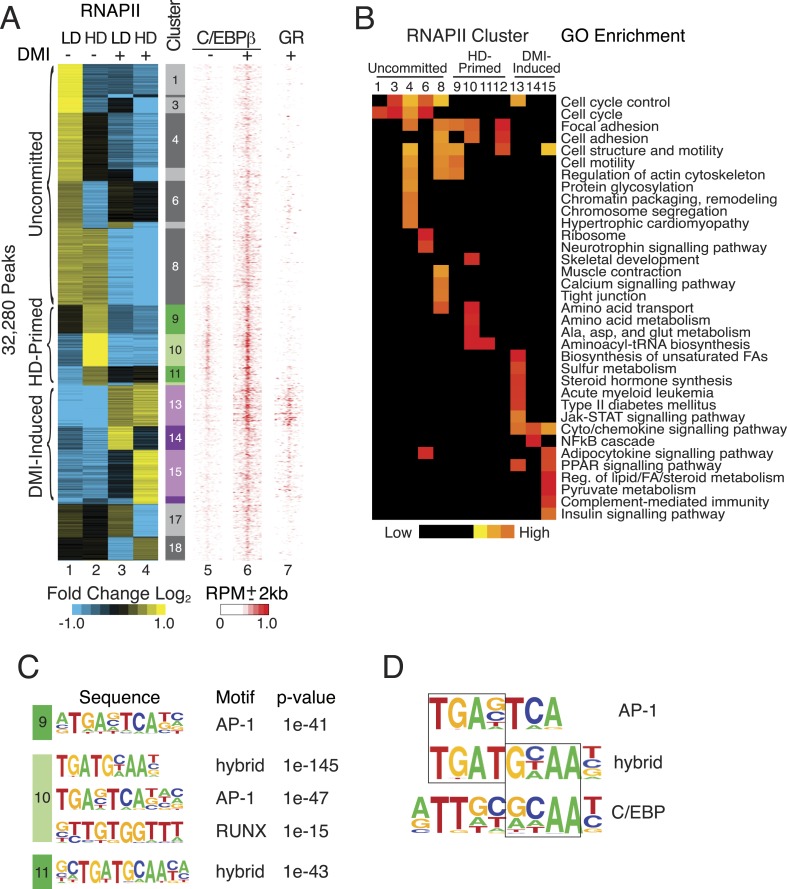
Figure 1. RNAPII-annotated enhancers reveal stage-specific transcriptional programs during adipogenic commitment.
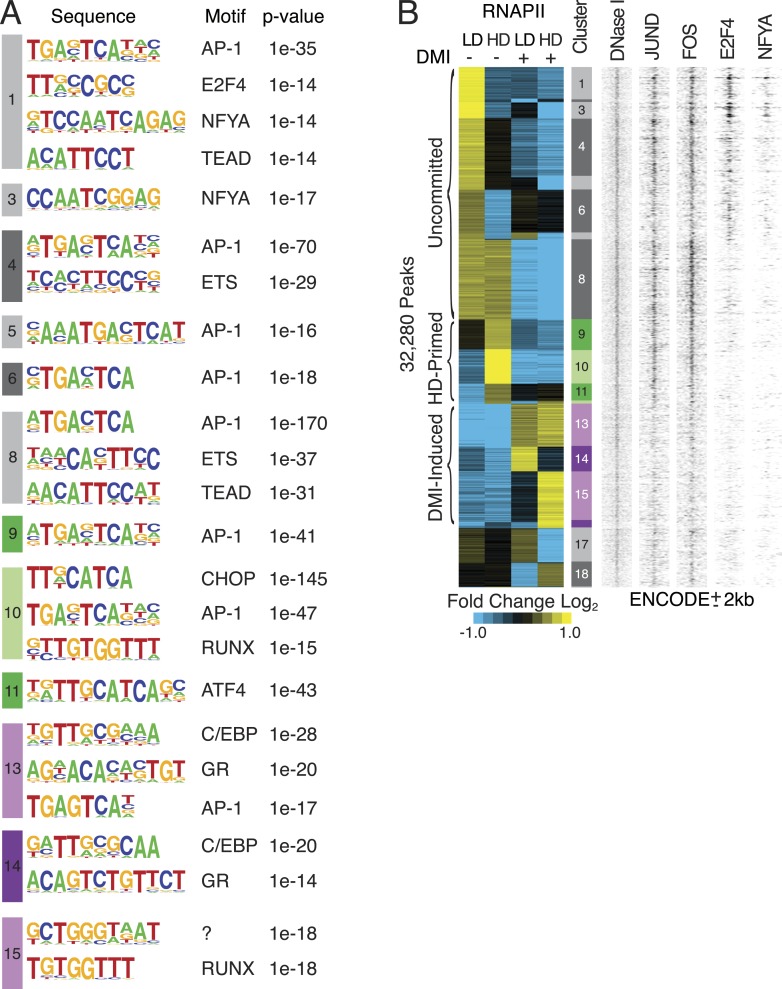
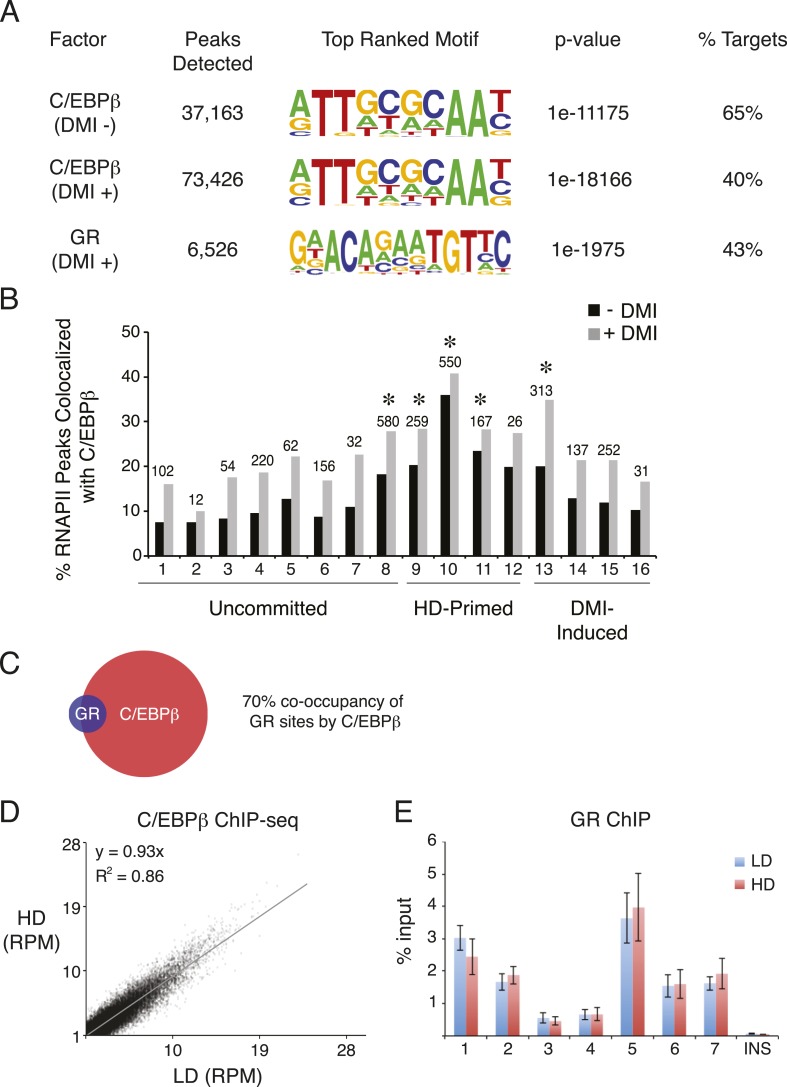
(A) Heat map of RNA Polymerase II (RNAPII) peaks clustered as a function of differential enrichment in response to changes in cell density (low, LD vs high, HD) and differentiation cocktail (+ or − DMI, 24 hr) in human mesenchymal stem cells (hMSCs) (tracks 1–4). Density heat maps (red) of C/EBPβ (0 and 6 hr DMI) and GR (6 hr DMI) binding in hMSCs within a 4 kb window of differential RNAPII peaks (tracks 5–7). Color bar specifically refers to scaling for C/EBPβ tracks. (B) Gene ontologies (GOs) resulting from mapping RNAPII enhancers to genes with correlated changes in gene body RNAPII. (C) Identification of de novo motifs in differential RNAPII clusters 9–11; reported sequences met detection thresholds of at least 5% of targets and p-value ≤ 1e-14. (D) Comparison of the AP-1, hybrid and C/EBP motifs, with the AP-1- and C/EBP-half sites boxed.