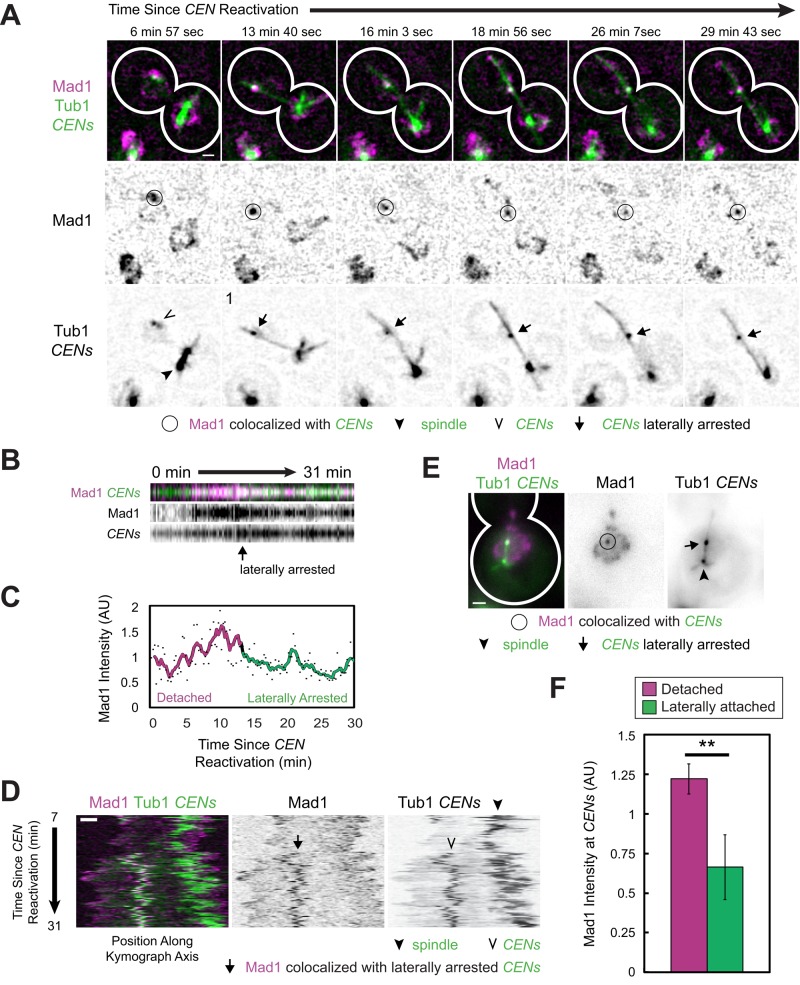
FIGURE 6:
Mad1 localization to sister centromeres associated laterally with microtubules. (A–D) kar3-64 cells expressing Mad1-3×mCherry, GFP-Tub1, and TetR-GFP and bearing MET3pr-CDC20 and GAL1pr-CEN3-TetOs were grown for 2.25 h at 25°C in YP medium plus 2 mM methionine, 2% raffinose, and 2% galactose to synchronize cells in metaphase and inactivate CEN3. The cells were shifted to 37°C, a restrictive temperature for both mutants, for 45 min, and then released into synthetic medium plus 2 mM methionine containing 2% glucose at 37°C. Cells were then examined by live-cell epifluorescence microscopy at 37°C, taking z-stacks every 14.4 s after centromere reactivation. Deconvolved, Gaussian-filtered MIPs are shown in A (Supplemental Video S7). (A) Time series of a representative cell in which the sister centromeres became laterally arrested. (B) Kymograph of a series of images approximately centered on the centromeres in A. (C) Mad1 intensity at the centromeres in A and B normalized by taking the ratio of the mCherry signal to the associated GFP intensity (dots). A color-coded sliding window average of five frames is also shown. (D) Kymograph along the axis connecting the spindle to the centromere after capture. (E, F) Cells of the same strain as in A–D were fixed after centromere reactivation at 15, 20, and 30 min before imaging by epifluorescence microscopy. (Data are from the same experiments as in Figure 3.) (E) Representative MIPs of laterally attached centromeres identified in fixed kar3-64 cells. (F) Normalized Mad1 intensity at detached and laterally attached sister centromeres. Mean of three independent experiments. Samples were 222, 292, and 315 detached centromeres and 16, 20, and 29 laterally attached centromeres, respectively. Error bars represent SEM. **p < 0.01. Scale bars, 1 μm.