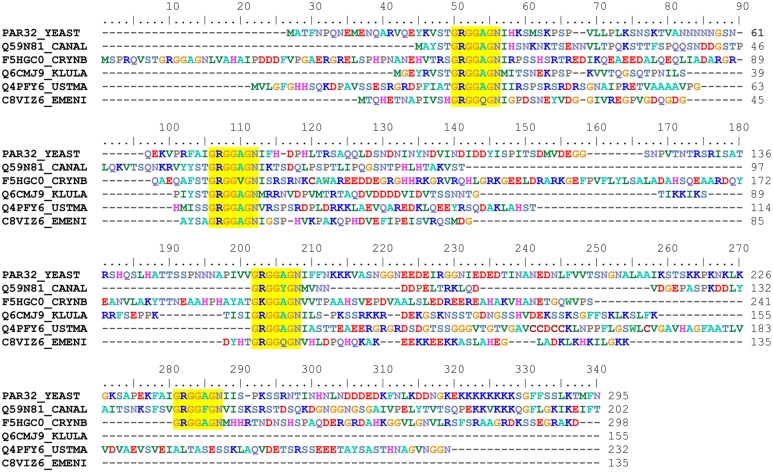
Fig 4. Amu1 is only conserved in fungi and contains a four-fold repeated motif.
Sequences were retrieved from the UniProt database [38] primarily by a BlastP search [39] and secondly by a FuzzPro search [40] using the following searching pattern, G-R-G-G-X-[AG]-N-X(30,110)-G-R-G-G-X-[AG]-N. A selection of Amu1/Par32 orthologues is displayed in the alignment; the aligned sequences being denoted by their accession numbers: PAR32_YEAST, Saccharomyces cerevisiae, Q59N81_CANAL, Candida albicans, F5HGC0_CRYNB, Crytococcus neoformans, Q6CMJ9_KLULA, Kluyveromyces lactis, Q4PFY6_USTMA, Ustilago maydis and C8VIZ6_EMENI, Aspergillus nidulans. The multiple sequence alignment was automatically generated by ClustalW [41] and manually adjusted using BioEdit [42]. The repeated motif characterizing the Amu1 protein family is highlighted in yellow.