Figure 1.
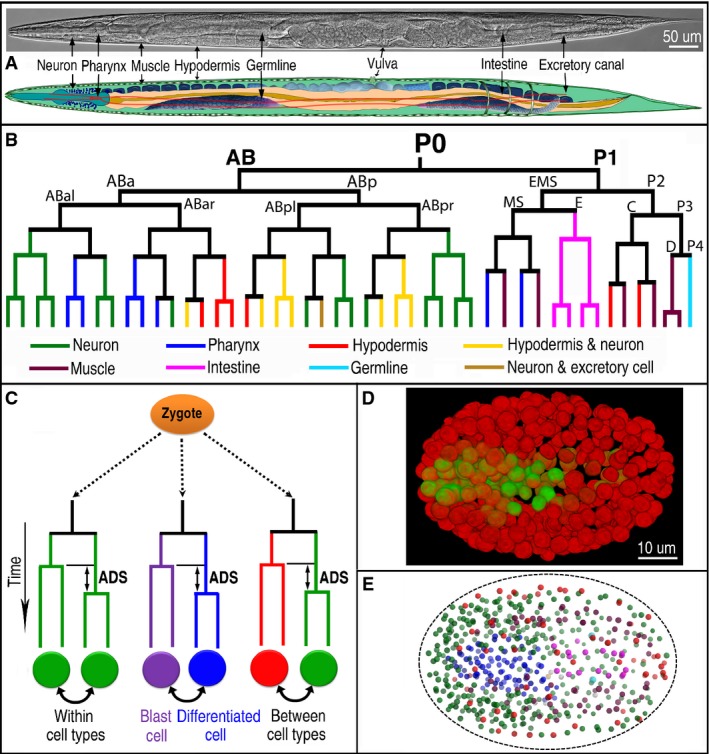
- A Nomarski micrograph (top) and a cartoon diagram (bottom) of a hermaphrodite adult showing major tissue types as indicated. Neuron, body-wall muscle, hypodermis, and excretory cell canal are not obvious in the Nomarski micrograph but are indicated based on their approximate positions.
- A lineage tree of an early C. elegans embryo (47 cells) showing various cell fates (differentially color coded) derived from different lineal origins.
- Schematic representation of ADS (asynchrony in cell division timing between sisters cells), which give rise to different or same cell type(s) as differentially color coded. Also shown is the comparison of asynchrony between the sister cells, one of which develops into a blast cell (purple) while the other becomes a terminally differentiated cell (blue) during embryogenesis.
- 3D projection of a C. elegans embryo of approximately 350-cell stage rendered with the fluorescence micrographs showing the expression of two lineaging markers, that is, pie-1::H2B::mCherry and H3.3::mCherry (red) and a pharynx-specific marker, PHA-4::GFP (green). See also Supplementary Movie S1 for expression dynamics and cell migrations.
- A reconstructed space-filling model of nuclei within a wild-type embryo of approximately 350-cell stage based on the output of automated lineaging. Nuclei are differentially color-coded based on their fates in the same way as that in (B). Dash line marks the approximate boundary of the embryo.
