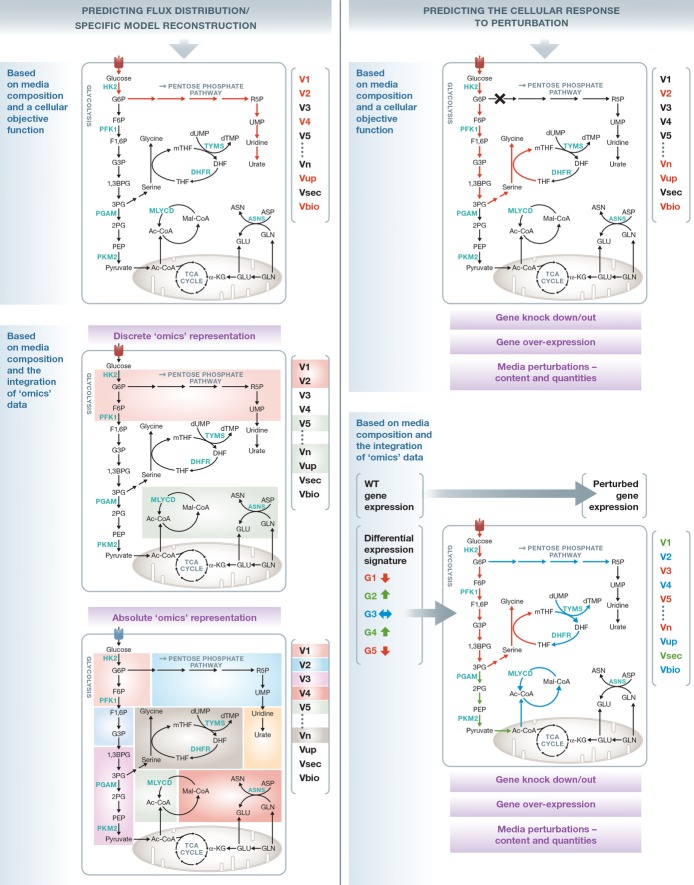
Figure 2.
Genome-scale metabolic modeling as a platform for predicting flux distributions and simulating cellular perturbations
Genome-scale metabolic modelings (GSMMs) provide an opportunity to characterize a cellular metabolic state by predicting the distribution of the network's reaction flux rates on a genome-scale level. For the analysis of microorganisms, this has been mostly achieved by assuming a pre-defined cellular objective function such as maximization of biomass yield or ATP production (left section, upper panel). Such an objective function cannot always be assumed when analyzing human metabolism, and therefore, omics data are utilized to derive a reduced specific model or characterize a metabolic flux state that best fits the context-specific omics data. The data can be used either in a discrete manner (left section, middle panel), trying to activate the flux thorough reactions associated with highly expressed genes (green) while removing those associated with lowly expressed genes (red), or constraining the model more quantitatively by considering the absolute expression levels (as depicted by the different colors, left section, lower panel). The network can be further studied by simulating genetic and environmental perturbations (right section). Similarly, the flux through the perturbed network can be derived based on a pre-defined objective function (right section, upper panel) or by utilizing the omics data to define the differential expression signature that can then be used to constrain the model in various ways (right section, lower panel).