Fig. 1. Telomeres and Telomerase.
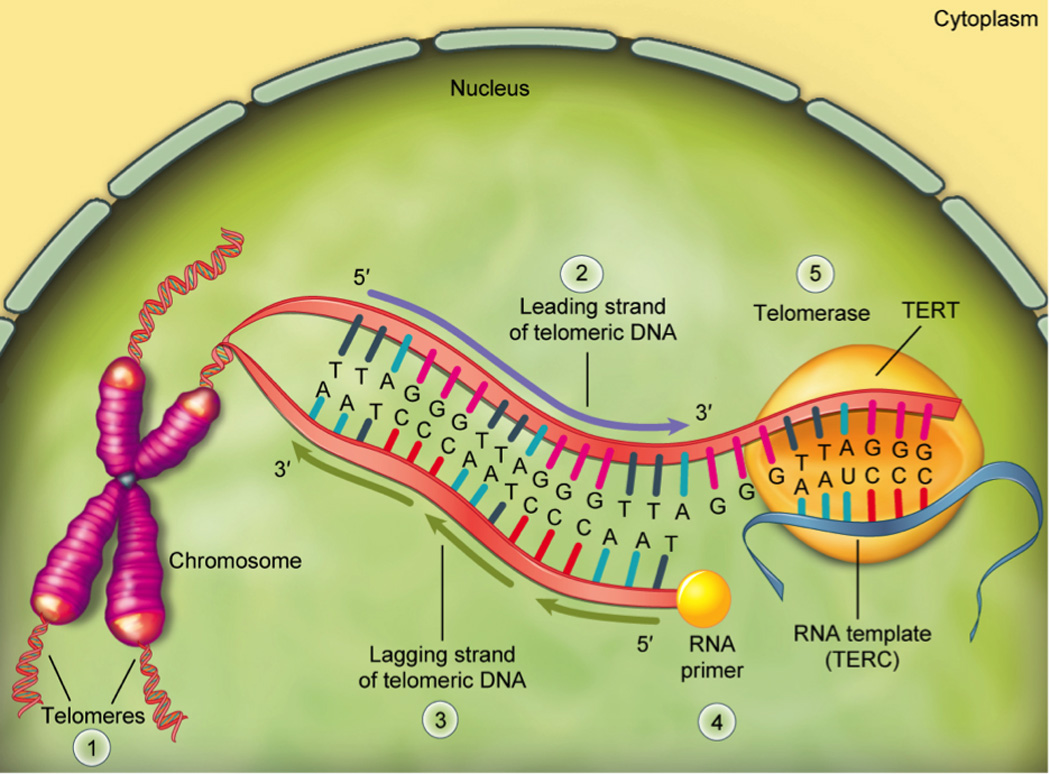
Telomeres [1] are protective caps at the ends of linear DNA strands. In humans, telomeres are comprised of multiple non-coding repeats of the nucleotide sequence, TTAGGG, and at birth, human telomeres are approximately 10,000 nucleotides long (Okuda et al., 2002). Telomeres lose approximately 50–100 nucleotides per DNA replication cycle (unless acted upon by telomerase) due to the so-called “end-replication problem” and can lose even more due to oxidative damage. The end replication problem arises during DNA replication or extension because DNA polymerase can only synthesize DNA in one direction (5’ → 3’). On the 5’ → 3’ leading strand [2], this route is continuous, but on the lagging strand [3], it is discontinuous, synthesized in fragments that require a RNA primer molecule [4] to provide a 5’ initiation point. As each fragment on the lagging strand (called “Okazaki fragments”) is completed, the RNA primer translocates to initiate the synthesis of additional fragments. Since the RNA primer must always attach prior to the synthesis of the lagging strand fragments, and since the RNA primer must base pair to complementary nucleotides on the leading strand, the 5’ end of lagging strand will always be shorter than the 3’ end of the leading strand, and thus is incompletely replicated. Shortened telomeres can be rebuilt by telomerase [5], which is comprised of the telomerase reverse transcriptase (TERT) enzyme and a telomerase RNA component (TERC) that serves as a template for new complementary telomeric DNA construction along the leading strand. As telomerase advances along the leading telomeric DNA strand, new nucleotides are added to it, providing additional room for extension of the lagging strand (Chakhparonian and Wellinger, 2003).
