Abstract
The trimethylguanosine (TMG) caps of small nuclear (sn) RNAs are synthesized by the enzyme Tgs1 via sequential methyl additions to the N2 atom of the m7G cap. Whereas TMG caps are inessential for Saccharomyces cerevisiae vegetative growth at 25° to 37°, tgs1∆ cells that lack TMG caps fail to thrive at 18°. The cold-sensitive defect correlates with ectopic stoichiometric association of nuclear cap-binding complex (CBC) with the residual m7G cap of the U1 snRNA and is suppressed fully by Cbc2 mutations that weaken cap binding. Here, we show that normal growth of tgs1∆ cells at 18° is also restored by a C-terminal deletion of 77 amino acids from the Snp1 subunit of yeast U1 snRNP. These results underscore the U1 snRNP as a focal point for TMG cap function in vivo. Casting a broader net, we conducted a dosage suppressor screen for genes that allowed survival of tgs1∆ cells at 18°. We thereby recovered RPO26 (encoding a shared subunit of all three nuclear RNA polymerases) and RPO31 (encoding the largest subunit of RNA polymerase III) as moderate and weak suppressors of tgs1∆ cold sensitivity, respectively. A structure-guided mutagenesis of Rpo26, using rpo26∆ complementation and tgs1∆ suppression as activity readouts, defined Rpo26-(78-155) as a minimized functional domain. Alanine scanning identified Glu89, Glu124, Arg135, and Arg136 as essential for rpo26∆ complementation. The E124A and R135A alleles retained tgs1∆ suppressor activity, thereby establishing a separation-of-function. These results illuminate the structure activity profile of an essential RNA polymerase component.
Keywords: trimethylguanosine synthase, U1 snRNP, RNA polymerase subunit, Rpo26
Trimethylguanosine (TMG) cap structures are characteristic of small nuclear (sn) RNAs, small nucleolar (sno) RNAs, and telomerase RNA. TMG is formed post-transcriptionally by the enzyme Tgs1, which catalyzes two successive methyl additions to the N2 atom of the m7G cap (Mouaikel et al. 2002; Hausmann and Shuman 2005; Chang et al. 2010). Whereas m7G caps are essential for the viability of eukaryal cells, TMG caps are not (Mouaikel et al. 2002; Hausmann et al. 2007). Saccharomyces cerevisiae and Schizosaccharomyces pombe tgs1∆ cells have no detectable TMG caps on their snRNAs, snoRNAs, or telomerase RNA, signifying that there is no Tgs1-independent route to form TMG caps (Mouaikel et al. 2002; Hausmann et al. 2007; Gallardo et al. 2008; Franke et al. 2008; Simoes-Barbosa et al. 2012). tgs1∆ yeast cells display apparently normal steady-state snRNA levels (Mouaikel et al. 2002; Hausmann et al. 2007) and no overt aberrations in the RNA or protein contents of their spliceosomal snRNPs, except for the acquisition of the nuclear cap-binding complex (CBC) as a stoichiometric component of the U1 snRNP (Schwer et al. 2011).
S. cerevisiae can grow in the absence of Tgs1 because the effects of ablating the TMG cap of the spliceosomal U snRNAs are genetically buffered, either by spliceosome assembly factors that are themselves inessential for vegetative growth (Hausmann et al. 2008; Wilmes et al. 2008; Chang et al. 2010) or by otherwise dispensable domains of the essential branchpoint binding protein Msl5 (Chang et al. 2012). Nonetheless, there are two situations in which the lack of TMG caps per se elicits a profound phenotype. First, S. cerevisiae tgs1∆ diploids are unable to properly execute meiosis and sporulation because they are defective in splicing certain meiotic pre-mRNAs (Qiu et al. 2011). Second, although S. cerevisiae haploid tgs1∆ cells grow normally at 30°–37°, they are unable to grow at 18°–20° (Mouaikel et al. 2002; Hausmann et al. 2008), signifying that one or more essential cellular transactions becomes reliant on TMG caps at low temperatures.
To gain insights to the basis for tgs1∆ cold sensitivity, we sought to identify genetic suppressors that restore the growth of tgs1∆ cells at restrictive temperature . Based on our findings that the residual m7G cap of the U1 snRNP in tgs1∆ cells is accessible to and occupied by nuclear CBC, we queried whether mutating the cap-binding site of CBC (in the Cbc2 subunit of yeast CBC) might suppress tgs1∆ cs growth. We thereby identified a series of hypomorphic mutations of Cbc2 predicted to weaken cap-binding (Mazza et al. 2002; Calero et al. 2002), which had no effect on vegetative growth in TGS1 cells yet restored the growth of tgs1∆ cells at 18°–20° (Schwer et al. 2011; Qiu et al. 2012) . We inferred from these results that the cs phenotype is caused, at least in part, by the ectopic association of nuclear CBP with the m7G cap of U1 snRNA.
This focused our attention on the U1 snRNP as a potential source of additional tgs1∆ suppressors. Yeast U1 snRNP consists of a 568-nt U1 snRNA, a 7-subunit Sm protein ring, and 10 U1-specific snRNP subunits: Prp39, Prp40, Snu71, Snu56, Luc7, Prp42, Nam8, Snp1, Mud1, and Yhc1 (Gottschalk et al. 1998; Schwer et al. 2011). The Nam8 and Mud1 subunits are inessential for vegetative growth. Rather than suppressing tgs1∆, we learned early on that nam8∆ and mud1∆ deletions were synthetic lethal with tgs1∆ (Hausmann et al. 2008). The essential Yhc1 subunit (the yeast homolog of human U1-C) interacts directly with the conserved U1 snRNA 5′ leader sequence m2,2,7GpppAUACUUACCU that base-pairs to the complementary sequence of the consensus yeast 5′ splice site (GUAUGU) (Kondo et al. 2015). Extensive mutagenesis of Yhc1, by C-terminal truncation and structure-guided alanine scanning, has yielded a collection of hypomorphic YHC1 alleles that have no effect per se on vegetative growth, but have strong negative genetic interactions with Mud1, Nam8, and Mud2 (Schwer and Shuman 2014, 2015). Testing the YHC1 mutant collection for interactions with tgs1∆ uncovered many instances of synthetic lethality and sickness, but no case in which the tgs1∆ cs phenotype was suppressed. The same was true of a series of truncation and alanine mutants of the SmD3 and SmB subunits of the yeast Sm ring (Schwer and Shuman 2015).
Despite these discouraging results, we continued the search for tgs1∆ suppressors along two lines, one U1-centric, and one unbiased. As we report here, both approaches bore fruit. In the first instance, we generated a series of viable truncation mutants of U1 subunit Snp1 and screened them for genetic interactions. We found two C-terminal truncations of Snp1 that restored the growth of tgs1∆ cell at low temperature.
In parallel, we screened a 2-µ plasmid-based yeast genomic library for candidate dosage suppressors of the tgs1∆ cs defect. The rationale was that bypass by overexpression might identify specific gene products or cellular transactions that are limiting when TMG caps are absent. We report the results of the screen, which identified the RNA polymerase subunit Rpo26 as capable of reviving the growth of tgs1∆ cells at restrictive temperature when expressed from plasmid vectors. Rpo26, a 155-aa polypeptide, is an essential constituent of all three nuclear RNA polymerases (Pol I, Pol II, and Pol III) (Archambault et al. 1990; Woychik et al. 1990). Rpo26 plays key roles in nuclear RNA polymerase assembly and function (Nouraini et al. 1996a; Tan et al. 2003). It makes direct atomic contacts to the catalytic Rpo21 subunit of yeast Pol II (Cramer et al. 2001), and hypomorphic mutations of Rpo26 are synthetic lethal in combination with an rpo21-ts allele (Archambault et al. 1990; Nouraini et al. 1996a). Rpo26 interacts similarly with the catalytic Rpa190 subunit in the crystal structure of yeast Pol I (Fernández-Tornero et al. 2013) and is presumed to do so with the catalytic Rpo31 subunit of Pol III. Our screen also identified Rpo31 as a weaker tgs1∆ suppressor.
We proceeded to conduct a structure-guided mutational analysis of Rpo26 and thereby delineated a minimal functional domain, Rpo26-(78-155), capable of rpo26∆ complementation and tgs1∆ cs suppression. We identified separation-of-function mutations within this domain that abolished rpo26∆ complementation without affecting tgs1∆ suppression.
Materials and Methods
Snp1 C-terminal truncations and tests of function
A 1.59-kbp DNA segment bearing the SNP1 gene (nucleotides −400 to +1190) was amplified from S. cerevisiae genomic DNA by PCR using primers that introduced restriction sites for inserting the gene into the yeast expression plasmids pRS316 (CEN URA3) and pRS413 (CEN HIS3). The resulting plasmids p316-SNP1 and p413-SNP1 were constructed to introduce a 5′ BamHI site and a 3′ SpeI site immediately flanking the open reading frame. C-terminal truncation alleles SNP1-(1-223), SNP1-(1-208), and SNP1-(1-193) were generated by PCR amplification with reverse primers that introduced a STOP codon in lieu of codons for Thr224, Ser209, or Phe194 and a flanking SpeI site. The mutated PCR fragments were digested and inserted into p413-SNP1 in lieu of the wild-type SNP1 gene. The plasmid-borne genes were sequenced completely to confirm that no unwanted changes were introduced during PCR and cloning.
To assess the effects of SNP1 mutations, we first generated a haploid snp1∆ [p316-SNP1] strain by sporulating and dissecting heterozygous SNP1snp1∆::kanMX diploids (Open Biosystems) that had been transfected with p316-SNP1. snp1∆ [p316-SNP1] cells were resistant to G418 and unable to grow on medium containing 5-fluoroorotic acid (FOA). To assess the function of SNP1 alleles, snp1∆ [p316-SNP1] cells were transfected with p413-SNP1 (HIS3 CEN) plasmids. His+ transformants were selected and streaked on agar medium containing FOA. The plates were incubated at 20°, 30°, and 37°, and mutants that failed to form colonies at any temperature after 8 d were deemed lethal. Individual FOA-resistant colonies of viable SNP1 alleles were grown to mid-log phase in YPD broth and adjusted to A600 of 0.1. Aliquots (3 µl) of serial 10-fold dilutions were spotted to YPD agar plates, which were then incubated at temperatures ranging from 18° to 37°. We also developed plasmid shuffle assays to test mutational effects on SNP1 function in tgs1∆, nam8∆, mud1∆, mud2∆, and CBC2-Y24A cells using standard genetic manipulations of mating, sporulation, and dissection.
Screen for dosage suppressors of tgs1Δ cold sensitivity
tgs1∆ cells were transfected with a yeast genomic DNA library in vector YEp24 (2 μ URA3). Approximately 41,000 Ura+ transformants were plated on medium lacking uracil at 18°. The 2 μ plasmid was isolated from 20 colonies that grew at 18° and then transformed into Escherichia coli. Plasmids were prepared from cultures of individual ampicillin-resistant transformants. The 20 candidate suppressor plasmids were re-tested by transformation into the tgs1∆ strain; 16 of them rescued growth of tgs1∆ cells at 18°. Primers flanking the cloning site were used to sequence the ends of the genomic DNA inserts in these 16 plasmids and thereby identify the genes contained in each clone. Eight of the clones contained the TGS1 gene. Six plasmids contained a yeast genomic DNA locus, provisionally named DTS1 (DTS = deletion of TGS1 suppressor). Two plasmids contained a different yeast genomic locus, provisionally named DTS2.
Yeast expression plasmids
The following DNA fragments with flanking BamHI sites at both 5′ and 3′ ends were amplified by PCR using DTS1 or DTS2 plasmids as templates: (i) the RPO26 ORF and intron (544 bp) plus 419 bp and 239 bp of 5′ and 3′ flanking genomic DNA; (ii) the MLC2 ORF (492 bp) plus 443 bp and 240 bp of 5′ and 3′ flanking genomic DNA; (iii) the PZF1 ORF (1.3 kbp) plus 455 bp and 244 bp of 5′ and 3′ flanking genomic DNA; (iv) the SKI3 ORF (3.0 kbp) plus 442 bp and 231 bp of 5′ and 3′ flanking genomic DNA; (v) the AZF1 ORF (2.7 kbp) plus 441 bp and 303 bp of 5′ and 3′ flanking genomic DNA; (vi) the TRS33 ORF plus 463 bp and 276 bp of 5′ and 3′ flanking genomic DNA; and (vii) the YOR114w ORF plus 475 bp and 232 bp of 5′ and 3′ flanking genomic DNA. A DNA fragment containing the RPO31 ORF (4.4 kbp) plus 426 bp of upstream (5′) and 230 bp of downstream (3′) chromosomal DNA was amplified by PCR from the DTS2 plasmid using primers that introduced SalI sites at both the 5′ and 3′ ends. The RPO26 intron was removed cleanly from its genomic fragment via two-stage overlap extension PCR to generate the cDNA (RPO26*) with its genomic DNA flanks and BamHI terminal restriction sites. The PCR products were then restricted at the terminal sites and inserted into yeast expression vector YEp24 (2 μ URA3). The restricted fragments of RPO26, RPO26*, and RPO31 were also inserted into yeast expression vector pRS415 (CEN LEU2). RPO26 was inserted into BamHI-cut pRS316 (CEN URA3) to yield pRS316-RPO26 for use in the plasmid shuffle assays described below. The plasmid inserts were sequenced completely to exclude the acquisition of unwanted changes during PCR amplification and cloning. Plasmids p360-TGS1 (CEN URA3TGS1) and pUN100-TGS1 (CEN LEU2TGS1) used as positive controls were described previously (Hausmann et al. 2008).
Rpo26 mutants
An intron-less RPO26 ORF was PCR-amplified with sense-strand primers designed to introduce a BamHI restriction site immediately upstream of the translation start codon and an antisense primer that introduced an XhoI site downstream of the stop codon. The PCR product was digested with BamHI and XhoI and then inserted into a yeast expression vector pRS425-TPI (2 μ LEU2) to yield pRS425-TPI-RPO26, in which expression of RPO26 is driven by the yeast TPI1 promoter, contained in a 2.2-kb PvuII fragment from pYX132 (Novagen).
Truncated RPO26 alleles were constructed by PCR amplification with: (i) sense strand primers that introduced a new start codon at the positions specified plus a flanking BamHI site and/or (ii) antisense strand primers that introduced a new stop codon after the positions specified plus a flanking XhoI site. Single-alanine mutations R79A, E89A, R97A, E124A, R135A, R136A, D145A, and E150A were introduced into RPO26-(78-155) by two-stage PCR overlap extension with mutagenic primer oligonucleotides. The PCR products containing the mutated RPO26 ORFs were digested with BamHI and XhoI and inserted into BamHI/XhoI-cut pRS425-TPI-RPO26 in lieu of the wild-type RPO26 gene. The inserts of all plasmid clones were sequenced to exclude the acquisition of unwanted mutations during amplification and cloning.
Plasmid shuffle assay for rpo26Δ complementation
A heterozygous diploid S. cerevisiae RPO26rpo26::kanMX heterozygous strain (purchased from Open Biosystems) was transformed with pRS316-RPO26. The resulting Ura+ diploid was sporulated and tetrads were dissected. We thereby recovered viable rpo26::kanMX haploids that were resistant to G418 and unable to grow on medium containing 0.75 mg/ml FOA (5-fluoroorotic acid), a drug that selects against the URA3RPO26 plasmid. The rpo26Δ strain was used to test plasmid-borne alleles of RPO26 for rpo26Δ complementation by plasmid shuffle as follows. rpo26Δ pRS316-RPO26 cells were transfected with pRS425-TPI-RPO26 plasmids containing wild-type or mutant RPO26 alleles. Individual transformants were selected and patched on SD-Leu agar medium. Cells from each isolate were streaked on agar medium containing 1.0 mg/ml FOA at 30°. In cases where the RPO26-containing plasmid supported growth on FOA, two isolates of each mutant amplified from single FOA-resistant colonies were tested for growth by spotting 3-μl aliquots of serial 10-fold dilutions of cells (from liquid cultures grown in SD–Leu medium to mid-log phase at 30° and adjusted to A600 of 0.1) to YPD agar and incubating the plates at 18° for 7 d, 25° for 4 d, 30° for 3 d, or 37° for 2 d.
Results
Synthetic genetics of Snp1 truncation mutants
Snp1, a 300-aa polypeptide, is the yeast homolog of human U1-70K (437-aa). Alignment of their primary structures highlights 98 positions of side-chain identity/similarity over the N-terminal 207-aa segment of Snp1 (Figure 1A). In the 3.3 Å crystal structure of the core human U1 snRNP (Kondo et al. 2015), the N-terminal 60-aa segment of U1-70K is a highly extended polypeptide that drapes across the surface of the U1 particle, making contacts to U1-C/Yhc1 near the U1 snRNA 5′ terminus, to each of the Sm ring subunits, and to the U1 snRNA 3′ of the Sm site. The segment of U1-70K from aa 61-202 (underlined in Figure 1A), comprising a long α helix and an RRM domain, binds to the conserved stem-loop 1 (SL1) of the U1 snRNA (Kondo et al. 2015). The C-terminal domains of Snp1 and U1-70K differ in length and amino acid sequence and are expected to be poorly structured based on their amino acid composition. The conserved N-terminal 21-aa peptide of Snp1 that interacts with U1-C/Yhc1 and SmD3 could be deleted without effect on yeast vegetative growth at any temperature (Schwer and Shuman 2015). A SNP1-(22-300) tgs1∆ double-mutant displayed the same cs growth defect as tgs1∆ (Schwer and Shuman 2015).
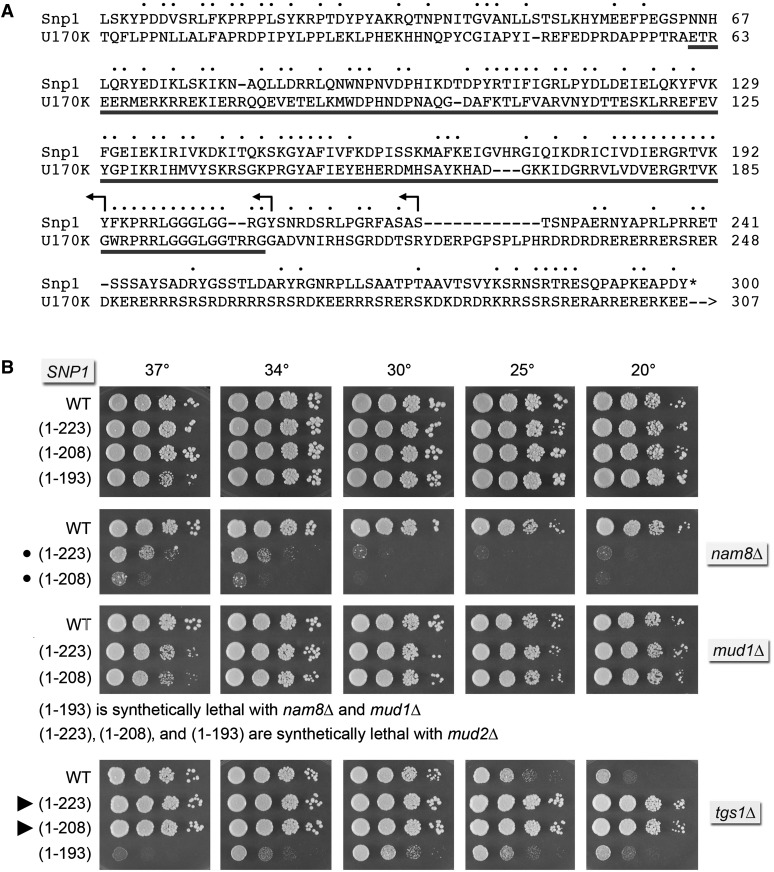
Figure 1.
Snp1 C-terminal truncations suppress tgs1∆ cold sensitivity. (A) The amino acid sequence of the 300-aa S. cerevisiae Snp1 protein is aligned to the homologous segment of the 437-aa human U1-70K polypeptide. Positions of side chain identity/similarity are indicated by • above the alignment. Arrowheads indicate the boundaries of the C-terminal truncations of Snp1. (B) The wild-type and truncated SNP1 alleles were tested for activity by plasmid shuffle in snp1∆, snp1∆ mud2∆, snp1∆ nam8∆, snp1∆ mud1∆, and snp1∆ tgs1∆ strains. Viable FOA-resistant snp1∆ strains bearing the indicated SNP1 allele on a CEN HIS3 plasmid in an otherwise wild-type (top panel), mud1∆, or nam8∆ background as indicated were spot-tested for growth on YPD agar at the temperatures specified. Synthetic growth defects are denoted by •. snp1∆ tgs1∆ strains bearing the indicated SNP1 allele on a CEN HIS3 plasmid were spot-tested for growth on YPD agar at the temperatures specified (bottom panel). Suppressors of the tgs1∆ cs phenotype are denoted by arrowheads.
Here, we constructed three C-terminal truncation mutants of Snp1 with distal margins indicated by the reverse arrowheads in Figure 1A. The wild-type and truncated alleles were placed on CEN HIS3 plasmids under the control of the native SNP1 promoter and tested by plasmid shuffle for complementation of a snp1∆ p[CEN URA3SNP1] strain. The resulting SNP1-(1-223), SNP1-(1-208), and SNP1-(1-193) strains were viable after FOA selection and grew as well as wild-type YHC1 cells on YPD agar (Figure 1B).
We surveyed genetic interactions of the benign Snp1 C-terminal truncations with mud2∆, nam8∆, and mud1∆. The results (Figure 1B) disclosed an informative hierarchy of synthetic mutational effects. SNP1-(1-223), SNP1-(1-208), and SNP1-(1-193) were lethal at all temperatures in the absence of Mud2, indicating that the essential contributions of the Snp1 segment downstream of the RRM module to early spliceosome assembly/stability are buffered by the cross-intron bridging interactions of Mud2 (engaged with Msl5 at the branchpoint) with U1 snRNP at the 5′ splice site.
SNP1-(1-223) and SNP1-(1-208) were barely viable in the nam8∆ genetic background and SNP1-(1-193) was synthetically lethal with nam8∆. By contrast, SNP1-(1-223) and SNP1-(1-208) supported normal growth of mud1∆ cells at 20°–34° and slightly slowed growth at 37° (Figure 1B). The salient finding was that the more truncated SNP1-(1-193) allele was synthetically lethal in the mud1∆ strain, signifying that the Snp1 peptide 194FKPRRLGGGLGGRGY208 is critical for U1 snRNP function in vivo in the absence of Mud1. The corresponding peptide in U1-70K makes direct contacts to the SL1 loop (Kondo et al. 2015).
SNP1-(1-223) and SNP1-(1-208) suppress tgs1Δ
The standout finding was that the SNP1-(1-223) and SNP1-(1-208) truncation alleles restored the growth of tgs1∆ cells at 25° and 20° (Figure 1B) and at 18° (not shown). Colony size of the SNP1-(1-223) tgs1∆ and SNP1-(1-208) tgs1∆ strains on YPD medium at cold temperatures was indistinguishable from the SNP1TGS1 wild-type strain (Figure 1B). Thus, deletion of the Snp1 segment from aa 224-300, downstream of the RRM domain, elicited a gain-of-function for the U1 snRNP that lacks a TMG cap. This positive genetic interaction was severed when the C-terminal truncation was extended into the RRM domain, i.e., the SNP1-(1-193) tgs1∆ reverted to cs growth (à la tgs1∆) and acquired a new ts phenotype (Figure 1B).
Mutational synergy of SNP1-(1-223) and SNP1-(1-208) with CBC2-Y24A
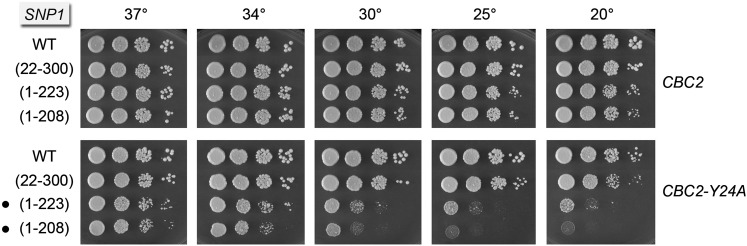
The Y24A mutation in the m7G-binding pocket of Cbc2 suppresses the tgs1∆ cs growth defect (Qiu et al. 2012), as do C-terminal truncations 1-223 and 1-208 of the U1 snRNP subunit Snp1. To query potential connections between Cbc2 and Snp1, we tested by plasmid shuffle the effects of the SNP1-(1-223) and SNP1-(1-208) alleles in CBC2snp1∆ and CBC2-Y24A snp1∆ strain backgrounds. We also tested in parallel the N-terminal truncation allele SNP1-(22-300), which eliminates a conserved peptide segment of Snp1/U1-70K that makes atomic contacts to the SmD3 and Yhc1/U1-C subunits of the U1 snRNP (Kondo et al. 2015; Schwer and Shuman 2015). Whereas SNP1-(22-300) caused no apparent growth defect in the CBC2-Y24A background, both SNP1-(1-223) and SNP1-(1-208) elicited a severe cold-sensitive defect in CBC2-Y24A cells (Figure 2), one that recapitulates the cold-sensitive growth defect of a cbc2∆ null strain (Qiu et al. 2012).
Figure 2.
Mutational synergy of SNP1-(1-223) and SNP1-(1-208) with CBC2-Y24A. The wild-type and truncated SNP1 alleles were tested for activity by plasmid shuffle in CBC2 snp1∆ and CBC2-Y24A snp1∆ strains. FOA-resistant isolates were spot-tested for growth on YPD agar at the temperatures specified. Synthetic growth defects are denoted by •.
RPO26 and RPO31 are dosage suppressors of tgs1Δ cs growth
The dosage suppressor screen entailed transformation of S. cerevisiae tgs1∆ cells with a 2-μ URA3 plasmid-based wild-type genomic DNA library and selection for Ura+ colonies that grew at 18°. Plasmid DNA was recovered from individual yeast colonies grown at 18° and then transformed into E. coli. Candidate suppressors were retransformed into the original tgs1∆ strain and tested for growth at 18°. Sequencing the insert junctions of the plasmids that retested faithfully revealed that the rescuing clones contained either TGS1 (as was to be expected) or one of two distinct extragenic suppressor loci, which we provisionally named DTS1 and DTS2, respectively (DTS = deletion of TGS1 suppressor). Note that whereas the 2-µ DTS1 or 2-µ DTS2 plasmids restored growth at 18°, compared to tgs1∆ cells carrying the empty 2-µ vector, neither 2-µ DTS1 nor 2-µ DTS2 was as effective as a TGS1 plasmid, as gauged by colony size (Figure 3).
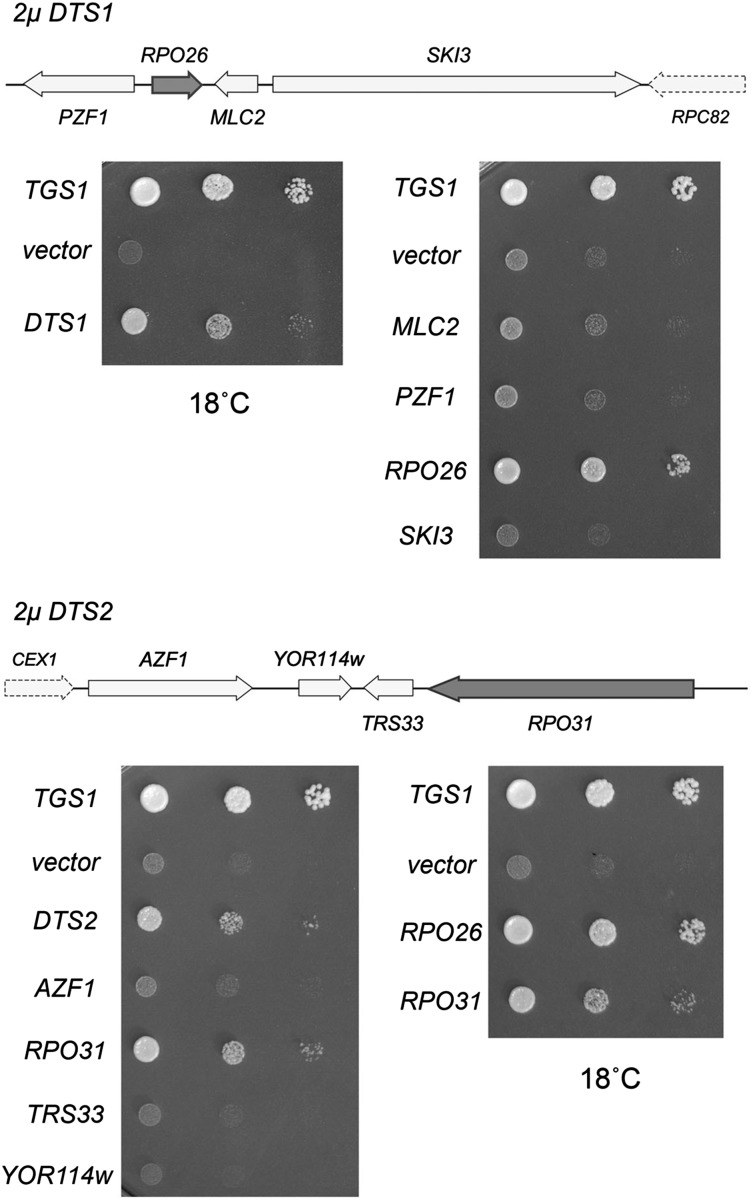
Figure 3.
RPO26 and RPO31 are dosage suppressors of tgs1Δ cold sensitivity. DTS1 (8.6 kb) and DTS2 (12.4 kb) are the two genomic inserts in the 2-μ URA3 plasmids that were isolated in the dosage suppressor screen for reversal of tgs1Δ cs growth. Individual genes with complete ORFs within the DTS1 and DTS2 inserts including PZF1, RPO26, MLC2, SKI3, AZF1, RPO31, TRS33, and YOR114w were cloned into a 2-μ URA3 vector. These plasmids, an empty CEN URA3 vector (negative control), a CEN URA3 plasmid containing wild-type TGS1 (positive control), and the plasmids containing DTS1 or DTS2 were transformed into tgs1Δ cells. Ura+ transformants were selected and grown at 30° in liquid medium lacking uracil. The cultures were adjusted to an A600 of 0.1 and aliquots of serial 10-fold dilutions were spotted on agar medium lacking uracil. The plates were photographed after incubation for 7 d at 18°.
Defining the suppressor loci within the 2µ DTS1 and DTS2 plasmids
DTS1 spans a segment of chromosome XVI that includes four complete genes—PZF1, RPO26, MLC2, and SKI3—plus a 3′ fragment of the RPC82 gene (Figure 3, top panel). To map the suppressor, we constructed a series of 2-µ vectors containing the individual PZF1, RPO26, MLC2, and SKI3 open reading frames and ∼200–400 bp of 5′ and 3′ flanking genomic DNA. tgs1∆ cells transformed with these plasmids were tested for growth at 18°, thereby revealing that the suppressor activity was inherent to RPO26 (Figure 3, top panel), the yeast gene encoding a shared 155-amino acid subunit of nuclear RNA polymerases I, II, and III.
DTS2 comprises a region of chromosome XV that embraces the complete AZF1, YOR114w, TRS33, and RPO31 genes, plus a 3′ fragment of the CEX1 gene (Figure 3, bottom panel). When 2-µ vectors containing the individual AZF1, YOR114w, TRS33, and RPO31 open reading frames and ∼200–400 bp of 5′ and 3′ flanking genomic DNA were transformed into tgs1∆ cells, only RPO31 revived growth at 18° compared to the vector control (Figure 3, bottom panel). Rpo31 encodes the 1460-amino acid largest subunit of nuclear RNA polymerase III. Side-by-side comparison of the growth of tgs1∆ cells bearing 2-µ RPO26 or RPO31 plasmids revealed that RPO26 was a better suppressor of the cold-sensitive defect, as gauged by colony size (Figure 3, bottom panel).
RPO26 and RPO31 suppress tgs1Δ at low gene dosage
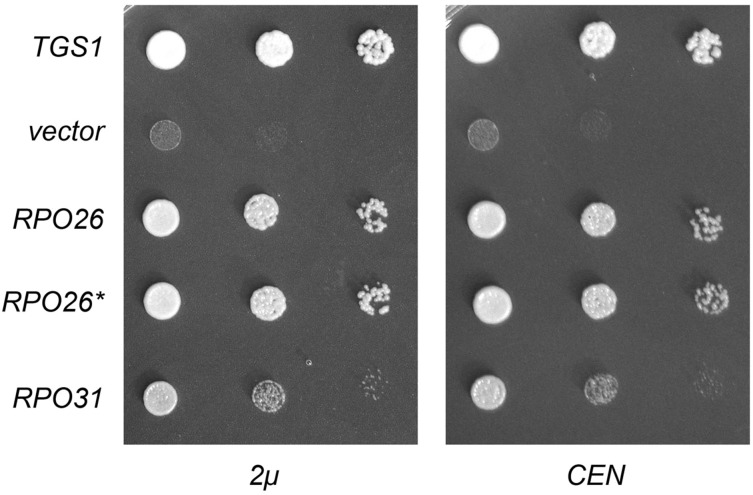
The identification of two RNA polymerase subunits as dosage suppressors of tgs1∆ suggested a novel connection between TMG caps and transcription. The connection via Rpo31 to RNA polymerase III, which is responsible for the synthesis of many essential noncoding RNAs (5S rRNA, U6 snRNA, tRNAs), was particularly puzzling insofar as none of the known Pol III transcripts have 5′ TMG (or m7G) caps. One scenario that might explain the genetic suppressor results is that the loss of TMG caps affects nucleolar architecture and function (Colau et al. 2004) such that the assembly or activity of Pol III is compromised at cold temperature, and this defect can be overcome, in part, by overexpressing either RPO31 or RPO26. If this is the case, then we might expect that simultaneously overexpressing RPO31 and RPO26 would afford better growth of tgs1∆ cells at 18° than increasing the gene dosage of either gene alone. We tested this by introducing RPO31 and RPO26 on the same 2-µ plasmid, but observed no better rescue of tgs1∆ growth in the cold than that afforded by 2-µ RPO26 (data not shown). Another prediction of the above scenario is that tgs1∆ suppression should require high gene dosage. To address this issue, we placed the RPO31 and RPO26 genes on CEN plasmids and transformed them into tgs1∆ cells. The striking finding was that provision of RPO26 or RPO31 on a CEN plasmid was just as effective as the 2-µ RPO26 or RPO31 plasmids in restoring tgs1∆ growth at restrictive temperature (Figure 4).
Figure 4.
RPO26 and RPO31, at low gene dosage, are capable of restoring growth of tgs1Δ at 18°. (Left) Yeast tgs1Δ cells were transformed with a CEN URA3 plasmid bearing wild-type TGS1 (positive control), an empty 2-μ URA3 vector (negative control), and 2-μ URA3 plasmids expressing wild-type RPO26, intron-less RPO26 cDNA (RPO26*), or RPO31. Ura+ transformants were selected at 30° and then tested for growth at 18° by spotting serial 10-fold dilutions of liquid cultures (grown at 30° in SD–Ura medium) on Ura− agar plates. The plates were photographed after incubation for 7 d at 18°. (Right) Yeast tgs1Δ cells were transformed with a CEN LEU2 plasmid bearing wild-type TGS1 (positive control), an empty CEN LEU2 vector (negative control), and CEN LEU2 plasmids expressing wild-type RPO26, RPO26*, or RPO31. Leu+ transformants were selected at 30° and then tested for growth at 18° by spotting serial 10-fold dilutions of liquid cultures (grown at 30° in SD-Leu medium) on SD-Leu agar plates. The plates were photographed after incubation for 7 d at 18°.
The aforementioned results point toward an alternative explanation for tgs1∆ suppression, whereby the lack of TMG caps selectively impacts the expression of RPO26 and/or RPO31, such that even one extra copy of these genes allows for growth in the cold. RPO26 seemed to us the more plausible target of such an effect, because: (i) TMG caps are certainly implicated genetically in pre-mRNA splicing; (ii) the RPO26 gene contains an intron, whereas RPO31 does not; and (iii) prior studies had shown that a 60% reduction in the level of mature RPO26 mRNA (caused by a mutation in the RPO26 promoter) resulted in a cold-sensitive growth defect (Nouraini et al. 1996b). We initially considered a scenario in which adequate Rpo26 expression might somehow require the presence of an intron in the pre-mRNA, akin to what has been described for the yeast Sus1 and the intron-containing SUS1 pre-mRNA (Cuenca-Bono et al. 2011; Hossain et al. 2011). In that case, we would expect that an intron-less cDNA version of RPO26 would not be able to suppress tgs1∆. However, we found that the RPO26 cDNA (designated RPO26* in Figure 3) was just as effective as the native RPO26 gene in promoting tgs1∆ growth at 18°, whether delivered on a 2-µ vector or a CEN vector (Figure 4).
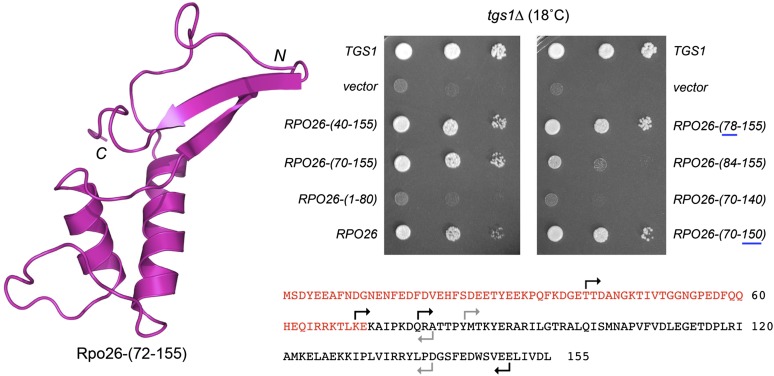
N- and C-terminal truncations of Rpo26 delineate a minimal functional domain
The crystal structure of yeast RNA polymerase II (Cramer et al. 2001) revealed the fold of the C-terminal segment of Rpo26 from amino acids 72 to 155, which comprises two α-helices and a β-hairpin (Figure 5). The N-terminal 71-amino-acid segment was disordered in the Pol II structure. In the recent crystal structure of yeast Pol I, the N-terminal 54-amino-acid segment of Rpo26 was disordered and the segment from amino acids 55 to 71 comprised an α-helix (Fernández-Tornero et al. 2013). A previous study had shown that deleting 42 amino acids from the N-terminus of Rpo26 did not affect the viability of yeast cells when the truncated RPO26-∆42 allele was driven by the strong GAL10 promoter in galactose-containing medium; however, deletion of 84 amino acids from the Rpo26 N-terminus was lethal (Nouraini et al. 1996a).
Figure 5.
N- and C-terminal truncations of Rpo26 delineate a minimal functional domain for tgs1∆ suppression. (Left) Tertiary structure of Rpo26-(72-155), from the yeast Pol II crystal structure (pdb 1I3Q), with the N and C termini indicated. (Bottom right) The amino acid sequence of yeast Rpo26. The C-terminal segment visualized in the Pol II crystal structure is in black font; the disordered N-terminal segment is in red font. The margins of the N- and C-terminal truncations are denoted by forward and reverse arrows. For the N-terminal deletions, the arrows specify the residues that were mutated to methionine to initiate the truncated proteins. Black arrows denote the truncations that allow the mutants to restore tgs1Δ growth at 18°, whereas the gray arrows denote the truncations that disable tgs1∆ suppression. (Top right) tgs1Δ cells were transformed with a CEN LEU2 plasmid bearing wild-type TGS1 (positive control), an empty 2-μ LEU2 TPI1 vector (negative control), or 2-μ LEU2 TPI1-RPO26 plasmids bearing wild-type RPO26 or the indicated truncation mutants. Leu+ transformants were selected at 30° and then tested for growth at 18° by spotting serial 10-fold dilutions of liquid cultures (grown at 30° in SD-Leu medium) on SD-Leu agar plates. The plates were photographed after incubation for 7 d at 18°.
Here, we tested the effects of finer incremental N- and C-terminal truncations on the in vivo activity of Rpo26, using two genetic readouts of function: (i) dosage suppression of tgs1∆ and (ii) complementation of rpo26∆. The truncated RPO26 alleles were placed on 2-µ plasmids under the control of the yeast TPI1 promoter. The N-terminal deletion alleles RPO26-(40-155), RPO26-(70-155), and RPO26-(78-155) were as effective as RPO26 in supporting tgs1∆ growth at 18°, whereas RPO26-(1-80), a truncated version encoding just the disordered N-terminal segment of Rpo26, had no salutary effect (Figure 5). The RPO26-(78-155) allele complemented rpo26∆ in a plasmid shuffle assay. RPO26-(78-155) cells grew as well as wild-type RPO26 yeast at 18°, 25°, 30°, and 37°, as gauged by colony size (Figure 6B). We conclude that the N-terminal 77 amino acids are dispensable for Rpo26 function as a subunit of the three nuclear RNA polymerases and as a suppressor of tgs1∆. By contrast, RPO26-(84-155) was a feeble suppressor of tgs1∆ at 18° (Figure 5) and was unable to complement rpo26∆ in the plasmid shuffle assay (not shown), signifying that the 78QRATTP83 peptide is important for Rpo26 activity.
Figure 6.
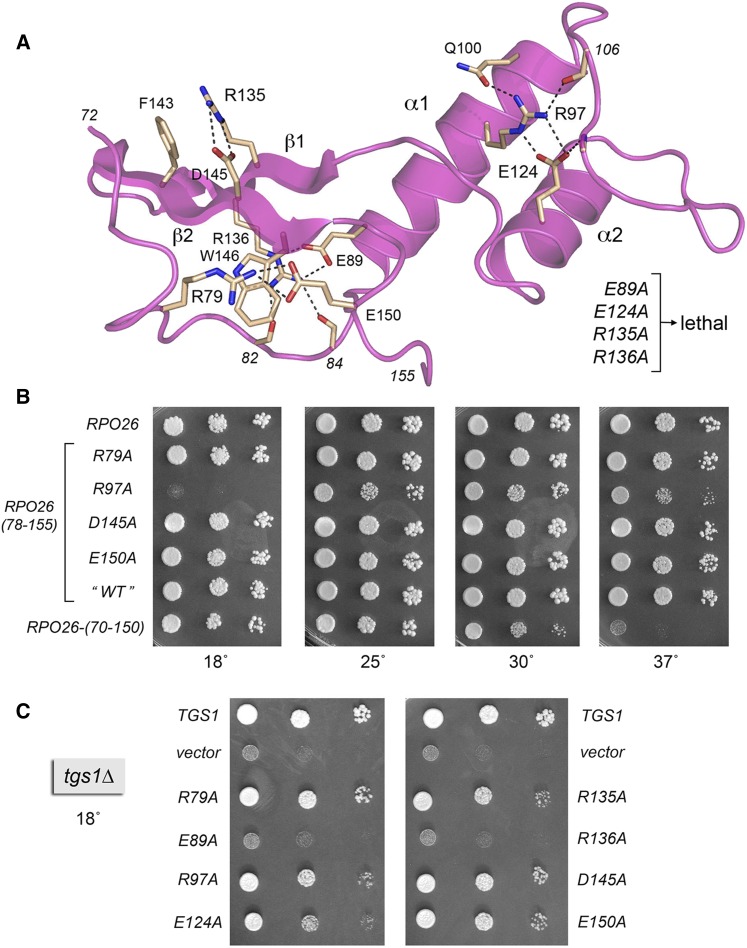
Structure-guided alanine scan of Rpo26. (A) Annotated structure of Rpo26 (from pdb 1I3Q) highlighting atomic interactions (dashed lines) of selected side chains and main chain atoms (depicted as stick models with beige carbons). Eight residues were targeted for alanine scanning: Arg79, Glu89, Arg97, Glu124, Arg135, Arg136, Asp145, and Glu150. The lethal RPO26-Ala alleles are indicated on the right. (B) rpo26∆ complementation. Yeast strain rpo26Δ p(URA3 CEN RPO26) was transformed with 2-μ LEU2 TPI1-RPO26 plasmids bearing wild-type RPO26, RPO26-(78-155), or the indicated RPO26-Ala mutants. Leu+ transformants were selected at 30° on agar medium containing 5-FOA (1.0 mg/ml) and aliquots of serial 10-fold dilutions of the strains with the specified genotypes were spotted on YPD agar medium. The plates were photographed after incubation for 2 d at 37°, 3 d at 30°, 5 d at 25°, or 7 d at 18°. (C) tgs1∆ suppression. tgs1Δ cells were transformed with a CEN LEU2 plasmid bearing wild-type TGS1 (positive control), an empty 2-μ LEU2 TPI1 vector (negative control), or 2-μ LEU2 TPI1-RPO26 plasmids bearing wild-type RPO26 or the indicated RPO26-Ala mutants. Leu+ transformants were selected at 30° and then tested for growth at 18° by spotting serial 10-fold dilutions of liquid cultures (grown at 30° in SD-Leu medium) on SD-Leu agar plates. The plates were photographed after incubation for 7 d at 18°.
We then tested the effects of deleting 5 amino acids or 16 amino acids from the C-terminus of the biologically active Rpo26-(70-155) polypeptide. Whereas RPO26-(70-150) was able to support growth of tgs1∆ cells at 18°, the RPO26-(70-140) allele was not (Figure 5). RPO26-(70-140) failed to complement rpo26∆ (not shown). By contrast, RPO26-(70-150) did complement rpo26∆, albeit with a conditional phenotype whereby RPO26-(70-150) cells grew well at 18° and 25°, formed small colonies at 30°, and failed to grow at 37° (Figure 6B). Thus, the decapeptide segment 141GSFEDWSVEE150 is necessary for Rpo21 function at warmer temperatures. Because a previous study had shown that a nonsense mutant allele encoding Rpo26-(1-145) was unable to complement rpo26∆ (Nouraini et al. 1996a), we can surmise that the pentapeptide 146WSVEE150 contains features essential for Rpo26 activity in vivo.
Structure-guided alanine scan identifies amino acids essential for rpo26Δ complementation
The crystal structure of Rpo26 in the context of RNA polymerase II highlights a network of intramolecular side chain contacts entailing salt bridge, hydrogen bonding, and π-cation interactions (Figure 6A). Here, we performed a structure-guided alanine scan of eight residues that comprise this network: Arg79, Glu89, Arg97, Glu124, Arg135, Arg136, Asp145, and Glu150. The alanine mutations were introduced into the biologically active RPO26-(78-155) gene on 2-µ plasmids under the control of the TPI1 promoter and tested for complementation of rpo26∆ by plasmid shuffle. Four of the alanine mutations were lethal: E89A, E124A, R135A, and R136A. Three of the alanine mutants—R79A, D145A and E150A—were viable and grew as well as “wild-type” RPO26-(78-155) at 18°, 25°, 30°, and 37° (Figure 6B). R97A cells grew at 25° and 30° but displayed cs and ts defects, whereby they failed to grow at 18° and grew slowly at 37°, as gauged by colony size (Figure 6B). We interpret the mutational data in light of the crystal structure, as follows.
Arg79 is located within the 78QRATTP83 hexapeptide defined as essential by our deletion analysis; Arg79 forms a salt bridge to Glu150 (Figure 6A), which is located within the essential C-terminal pentapeptide 146WSVEE150. Yet alanine mutation of either Arg79 or Glu150 was benign in vivo (Figure 6B), signifying that this salt bridge is dispensable and that one or more other constituents of the 78QRATTP83 and 146WSVEE150 peptides must be essential for Rpo26 function. In the case of the proximal peptide segment, we suspect that the key contributions are the hydrogen bonds of the main-chain Thr82 and Tyr84 carbonyls to the terminal guanidinium nitrogens of the essential Arg136 side chain (Figure 6A). For the distal 146WSVEE150 peptide, the Trp146 side chain is the likely key constituent, insofar as Trp146 is the focus of an extensive interaction network; it forms a cation–π–cation sandwich between Arg79 and Arg136 (Figure 6A) and it donates a hydrogen bond from Nε to the Glu144-Oε1 atom.
The essential Glu89 side chain, located in the first α-helix, forms a bidentate ion pair to the Nε and NH2 atoms of the essential Arg136 side chain, which is situated in the first β-strand (Figure 6A). Glu89 also receives a hydrogen bond to Oε1 from the main-chain amide of Thr86. We surmise that the Glu89-Arg136 salt bridge and the atomic contacts that Glu89 and Arg136 make to the main-chain of the 82TPYMT86 peptide loop preceding the first α-helix are necessary for Rpo26 folding and function. A conservative R136K mutation in RPO26 elicits a temperature-sensitive growth defect (Nouraini et al. 1996a).
Arg135 and Asp145 are situated on the opposite face of the β-hairpin, where they form an interstrand salt bridge (Figure 6A). It was noteworthy that whereas subtracting the Asp145 side chain had no apparent impact on cell growth, the loss of Arg135 was lethal. Thus, the Asp-Arg salt bridge is not essential for Rpo26 activity. Arg135 forms a cation-π stack on Phe143 (Figure 6A), and we suspect that this cation-π interaction accounts for the essentiality of Arg135. Consistent with this idea, replacing Arg135 with lysine, which would, in principle, preserve the cation-π interaction, had no effect on yeast growth (Nouraini et al. 1996a).
The essential Glu124 side chain participates in a network of ionic and hydrogen bond contacts involving the two α-helices and the connecting loop (Figure 6A). Glu124 (in α1) makes a bidentate salt bridge to Arg97 (in α1 and conditionally essential at 18°) and accepts a hydrogen bond from the main-chain amide of Phe108 (in the loop). Arg97, in turn, donates hydrogen bonds to Gln100 and the main-chain carbonyl of Pro106. It was shown previously that replacing Gln110 with arginine results in cold-sensitive and temperature-sensitive growth defects (Tan et al. 2003).
Rpo26 mutations that separate rpo26Δ complementation and tgs1Δ suppression activities
As one might expect, the R79A, D145A, and E150A mutants that complemented rpo26∆ at 18° were also active in suppressing tgs1∆ (Figure 6C). Mutations E89A and R136A that unconditionally abolished rpo26∆ complementation also eliminated tgs1∆ suppression. The salient findings were that: (i) two other mutants, R135A and E124A, that were unconditionally defective in rpo26∆ complementation retained tgs1∆ suppressor activity and (ii) mutant R97A, which was inactive in rpo26∆ complementation at 18°, nonetheless complemented tgs1∆ growth at 18°. Thus, R135A, E124A, and R97A exemplify separation of function mutations that distinguish the global role of Rpo26 in transcription by all nuclear RNA polymerases from its particular ability to act as a dosage suppressor of the cold sensitivity of tgs1∆ cells.
Discussion
The present study provides new genetic insights to the impact of the lack of TMG caps in budding yeast. Prior screening for synthetic lethal and sick tgs1∆ interactions had drawn attention specifically to the U1 snRNP as a focal point for TMG cap function in vivo (Hausmann et al. 2008). This idea was fortified by the findings that the only overt change in the composition of yeast spliceosomal snRNPs in tgs1∆ cells was the gain of CBC as a stoichiometric component of the U1 snRNP, by virtue of its binding to the residual m7G cap on the U1 snRNA (Schwer et al. 2011). In TGS1 cells, CBC is loosely associated with the U1 snRNP at low sub-stoichiometric levels compared to the intrinsic U1 snRNP subunits (Schwer et al. 2011). It is thought that CBC interacts with one or more of the U1 snRNP proteins to facilitate bridging interactions between CBC bound to the pre-mRNA m7G cap and the U1 snRNP at the 5′ splice site (Lewis et al. 1996; Görnemann et al. 2005). Hypomorphic mutations in Cbc2 that weaken cap binding suppress the tgs1∆ cs growth defect (Qiu et al. 2012).
Here, we show that restoration of growth of tgs1∆ cells in the cold can also be achieved by deleting the C-terminal 77-aa segment of the essential Snp1 subunit of the U1 snRNP. Although it had been appreciated earlier that this C-terminal portion of Snp1 was dispensable for vegetative growth (Hilleren et al. 1995), the genetic interactions of the Snp1-C∆ truncations were not interrogated. Underscoring the theme of redundancy in the yeast U1 snRNP, we show that otherwise benign Snp1-(1-223) and Snp1-(1-208) mutations are catastrophic in the absence of Mud2 or Nam8. Yet these same SNP1 truncation alleles elicit a gain-of-function in the tgs1∆ genetic background. Our frugal speculation is that tgs1∆ suppression by SNP1-C∆ is mediated via an effect on CBC association with the residual U1 snRNA m7G cap, whereby the C-terminal segment of Snp1 is itself a point of contact of CBC with the U1 snRNP. In this scenario, weakening of the CBC•U1 snRNP interaction by Snp1 truncation would diminish CBC association with the U1 m7G cap in tgs1∆ cells and allow growth in the cold (more or less mimicking the Cbc2 cap-binding site mutants with respect to tgs1∆ suppression). In TGS1 cells that have TMG caps, the Cbc2 cap-binding site lesion Y24A and the Snp1-C∆ truncations, which cause no growth defects per se, synergized when combined to mimic the severe cold sensitivity of the cbc2∆ null mutant. These findings are consistent with the idea that the Snp1 C-terminus contributes to the interaction of CBC with the U1 snRNP.
The C-terminal 77-aa segment of yeast Snp1 is rich in arginine (n = 11), serine (n = 12), and alanine (n = 11) and is predicted to be strongly hydrophilic, with the exception of one hydrophobic tract (265PLLSAATPTAAVTSVY280). The amino acid sequence and composition are suggestive of structural disorder or a structure that is templated by the association of this polypeptide with other proteins. Because this segment is not conserved in human U1-70K and the C-terminus of U1-70K is not seen in U1 snRNP crystal structures, we cannot intuit what contacts might be made by the Snp1 C-terminus. This will be an interesting topic for future studies given the broad impact, both positive and negative, of Snp1 C-terminal deletion on yeast physiology when other components of the splicing apparatus are simultaneously perturbed.
In a separate approach entailing a genomic library screen, we identified RPO26 as a dosage suppressor of the cold-sensitive phenotype of tgs1∆ cells. Because even a nominally single extra copy of the RPO26 gene on a CEN plasmid revived tgs1∆ growth at 18°, we surmise that RPO26 is an especially vulnerable target of the effect of the tgs1∆ mutation. This vulnerability is unlikely to reflect a fastidious gene-specific requirement for TMG caps or other splicing factors in removing the RPO26 intron, insofar as the RPO26 5′-splice site, branchpoint, and 3′-splice site adhere perfectly to the yeast consensus sequences and the intron is situated close to the 5′ end of the RPO26 ORF, as is the case for most yeast genes. Rather, it is the fact that even small changes in RPO26 expression can result in an overt growth defect in the cold (Nouraini et al. 1996b) that allowed us to recover a singularly sensitive target gene in the suppressor screen.
As discussed above, we implicate ectopic binding of nuclear CBC to the m7G cap of the U1 snRNP of tgs1∆ cells as a principal factor in the cold sensitivity of tgs1∆ cells. Mutations in the cap-binding site of CBC or deletion of the Snp1 C-terminus completely restore normal growth of tgs1∆ cells at 18°, unlike the dosage suppression by RPO26, which promotes growth of tgs1∆ cells at 18°, albeit not as well as TGS1 or the hypomorphic CBC2 and SNP1-C∆ mutations. These findings fortify the inference that the cold sensitivity of tgs1∆ arises not from the lack of TMG caps, but from the effect of U1-bound CBC on vulnerable yeast mRNAs, among which RPO26 stands out.
Our studies shed new light on the structure-activity relations of Rpo26. We refine the margins of the minimal functional Rpo26 domain, identify essential side chains by alanine scanning, and interpret the mutational effects by reference to the crystal structure of Rpo26 in RNA polymerase II. Especially instructive were the mutations that separated the tgs1∆ dosage suppression activity of Rpo26 from its globally essential function in nuclear transcription. An appealing explanation for this separation is that certain Rpo26 mutations selectively impact Rpo26 function in one (or two) of the nuclear RNA polymerases while sparing its function in the other polymerase(s) (Tan et al. 2003). In that case, we infer that the RPO26 R135A, E124A, and R97A mutants, which are lethal or conditionally lethal with respect to rpo26∆ complementation, can provide Rpo26 function for the nuclear RNA polymerase that is most affected in tgs1∆ cells at low temperatures. That we isolated RPO31, the gene encoding the largest subunit of Pol III, in the same suppressor screen that yielded RPO26 suggests to us that Pol III is especially sensitive to the level of Rpo26 subunit in tgs1∆ cells at low temperatures. Rpo26 is in intimate contact with the large subunits of nuclear RNA polymerases during and after assembly of the polymerases (Wild and Cramer 2012) and there are well-documented genetic interactions of Rpo26 with Rpb1, the largest Pol II subunit (Archambault et al. 1990; Nouraini et al. 1996a). We speculate that the relatively weaker suppression of tgs1∆ by increased RPO31 gene dosage (compared to RPO26 suppression) reflects enhanced assembly of Pol III when Rpo26 levels are limiting.
Acknowledgments
This work was supported by U.S. National Institutes of Health grants GM52470 (S.S.) and GM102961 (B.S.). S.S. is an American Cancer Society Research Professor.
Footnotes
Communicating editor: J. Rine
Literature Cited
- Archambault J., Schappert K. T., Friesen J. T., 1990. A suppressor of an RNA polymerase II mutations of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II and III. Mol. Cell. Biol. 10: 6123–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero G., Wilson K. F., Ly T., Rios-Steiner J. L., Clardy J. C., et al. , 2002. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 9: 912–917. [DOI] [PubMed] [Google Scholar]
- Chang J., Schwer B., Shuman S., 2010. Mutational analyses of trimethylguanosine synthase (Tgs1) and Mud2: proteins implicated in pre-mRNA splicing. RNA 16: 1018–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Schwer B., Shuman S., 2012. Structure-function analysis and genetic interactions of the yeast branchpoint binding protein Msl5. Nucleic Acids Res. 40: 4539–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colau G., Thiry M., Leduc V., Bordonné R., Lafontaine D. L. J., 2004. The small nucle(ol)ar RNA cap trimethyltransferase is required for ribosome synthesis and intact nuclear morphology. Mol. Cell. Biol. 24: 7976–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Bushnell D. A., Kornberg R. D., 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292: 1863–1876. [DOI] [PubMed] [Google Scholar]
- Cuenca-Bono B., García-Molinero V., Pascual-García P., Dopazo H., Llopis A., et al. , 2011. SUS1 introns are required for efficient mRNA nuclear export in yeast. Nucleic Acids Res. 39: 8599–8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Tornero C., Moreno-Morcillo M., Rashid U. J., Taylor N. M. I., Ruiz F. M., et al. , 2013. Crystal structure of the 14-subunit RNA polymerase I. Nature 502: 644–649. [DOI] [PubMed] [Google Scholar]
- Franke J., Gehlen J., Ehrenhofer-Murray A. E., 2008. Hypermethylation of yeast telomerase RNA by the snRNA and snoRNA methyltransferase Tgs1. J. Cell Sci. 121: 3553–3560. [DOI] [PubMed] [Google Scholar]
- Gallardo F., Olivier C., Dandjinoud A. T., Welinger R. J., Chartrand P., 2008. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 27: 748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görnemann J., Kotovic K. M., Hujer K., Neugebauer K. M., 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell 19: 53–63. [DOI] [PubMed] [Google Scholar]
- Gottschalk A., Tang J., Puig O., Salgado J., Neubauer G., et al. , 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393. [PMC free article] [PubMed] [Google Scholar]
- Hausmann S., Shuman S., 2005. Specificity and mechanism of RNA cap guanine-N2 methyltransferase (Tgs1). J. Biol. Chem. 280: 4021–4024. [DOI] [PubMed] [Google Scholar]
- Hausmann S., Ramirez A., Schneider S., Schwer B., Shuman S., 2007. Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe. Nucleic Acids Res. 35: 1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S., Zheng S., Costanzo M., Brost R. L., Garcin D., et al. , 2008. Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of TMG caps, snRNP components, pre-mRNA splicing factors, and RNA decay pathways. J. Biol. Chem. 283: 31706–31718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. J., Kao H. Y., Siliciano P. G., 1995. The amino-terminal domain of yeast U1–70K is necessary and sufficient for function. Mol. Cell. Biol. 15: 6341–6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M. A., Rodriguez C. M., Johnson T. L., 2011. Key features of the two-intron Saccharomyces cerevisiae gene SUS1 contribute to its alternative splicing. Nucleic Acids Res. 39: 8612–8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Oubridge C., van Roon A. M., Nagai K., 2015. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. eLife 4: e04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Izaurralde E., Jarmolowsk A., McGuinan C., Mattaj I. W., 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 10: 1683–1698. [DOI] [PubMed] [Google Scholar]
- Mazza C., Segref A., Mattaj I. W., Cusack S., 2002. Large-scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21: 5548–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel J., Verheggen C., Bertrand E., Tazi J., Bordonné R., 2002. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 9: 891–901. [DOI] [PubMed] [Google Scholar]
- Nouraini S., Archambault J., Friesen J. D., 1996. a Rpo26p, a subunit common to yeast RNA polymerases, is essential for the assembly of RNA polymerases I and II and for the stability of the largest subunits of these enzymes. Mol. Cell. Biol. 16: 5985–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouraini S., Hu J., McBroom L. D. B., Friesen J. D., 1996. b Mutations in an Abf1p binding site in the proximal promoter of yeast RPO26 shift the transcription start sites and reduced the level of RPO26 mRNA. Yeast 12: 1339–1350. [DOI] [PubMed] [Google Scholar]
- Qiu Z. R., Shuman S., Schwer B., 2011. An essential role for trimethylguanosine RNA caps in Saccharomyces cerevisiae meiosis and their requirement for splicing of SAE3 and PCH2 meiotic pre-mRNAs. Nucleic Acids Res. 39: 5633–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z. R., Chico L., Chang J., Shuman S., Schwer B., 2012. Genetic interactions of hypomorphic mutations in the m7G cap binding pocket of yeast nuclear cap binding complex: an essential role for Cbc2 in meiosis via splicing of MER3 pre-mRNA. RNA 18: 1996–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Erdjument-Bromage H., Shuman S., 2011. Composition of yeast snRNPs and snoRNPs in the absence of trimethylguanosine caps reveals nuclear cap binding protein as a gained U1 component implicated in the cold-sensitivity of tgs1∆ cells. Nucleic Acids Res. 39: 6715–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Shuman S., 2014. Structure-function analysis of the Yhc1 subunit of yeast U1 snRNP and genetic interactions of Yhc1 with Mud2, Nam8, Mud1, Tgs1, U1 snRNA, SmD3 and Prp28. Nucleic Acids Res. 42: 4697–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Shuman S., 2015. Structure-function analysis and genetic interactions of the Yhc1, SmD3, SmB, and Snp1 subunits of yeast U1 snRNP and genetic interactions of SmD3 with U2 snRNP subunit Lea1. RNA (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Barbosa A., Chakrabarti K., Pearson M., Benarroch D., Shuman S., et al. , 2012. Box H/ACA snoRNAs are preferred substrates for the trimethylguanosine synthase in the divergent unicellular eukaryote Trichomonas vaginalis. RNA 18: 1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q., Prysak M. H., Woychik N. A., 2003. Loss of the Rpb4/Rpb7 subcomplex in a mutant form of the Rpb6 subunit shared by RNA polymerases I, II, and III. Mol. Cell. Biol. 23: 3329–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T., Cramer P., 2012. Biogenesis of multisubunit RNA polymerases. Trends Biochem. Sci. 37: 99–105. [DOI] [PubMed] [Google Scholar]
- Wilmes G. M., Bergkessel M., Bandyopadhyay S., Shales M., Braberg H., et al. , 2008. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell 32: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N. A., Liao S. M., Kolodziej P. A., Young R. A., 1990. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev. 4: 313–323. [DOI] [PubMed] [Google Scholar]