Highlights
-
•
We have constructed Z. tritici ku70 and ku80 null mutants.
-
•
Gene targeting frequency in the ku null strains is greater than 85%.
-
•
Deletion of KU70 and KU80 does not affect in vitro growth or pathogenicity.
-
•
Sulfonylurea resistance was established as a new positive selection marker in Z. tritici.
-
•
Ternary vectors were constructed to enable yeast recombinational cloning in Z. tritici.
Keywords: Non homologous end joining mutants, Ku70, Ku80, Homologous recombination, Gene targeting frequency, Yeast recombinational cloning, Agrobacterium tumefaciens mediated transformation, Sulfonylurea, Zymoseptoria tritici
Abstract
The lack of techniques for rapid assembly of gene deletion vectors, paucity of selectable marker genes available for genetic manipulation and low frequency of homologous recombination are major constraints in construction of gene deletion mutants in Zymoseptoria tritici. To address these issues, we have constructed ternary vectors for Agrobacterium tumefaciens mediated transformation of Z. tritici, which enable the single step assembly of multiple fragments via yeast recombinational cloning. The sulfonylurea resistance gene, which is a mutated allele of the Magnaporthe oryzae ILV2 gene, was established as a new dominant selectable marker for Z. tritici. To increase the frequency of homologous recombination, we have constructed Z. tritici strains deficient in the non-homologous end joining pathway of DNA double stranded break repair by inactivating the KU70 and KU80 genes. Targeted gene deletion frequency increased to more than 85% in both Z. tritici ku70 and ku80 null strains, compared to ⩽10% seen in the wild type parental strain IPO323. The in vitro growth and in planta pathogenicity of the Z. tritici ku70 and ku80 null strains were comparable to strain IPO323. Together these molecular tools add significantly to the platform available for genomic analysis through targeted gene deletion or promoter replacements and will facilitate large-scale functional characterization projects in Z. tritici.
1. Introduction
The ascomycete fungal pathogen Zymoseptoria tritici causes Septoria tritici leaf blotch (STB) in cultivated wheat (Triticum species). It is one of the most economically important pathogens of wheat in the Northern Hemisphere and can reduce annual wheat yields by up to 50% if the disease is not controlled (Polley and Thomas, 1991). The pathogen overwinters as ascospores, which are dispersed by wind to initiate polycyclic STB infection in susceptible hosts (Orton et al., 2011). Z. tritici ascospores germinate on leaf surfaces and invade the host through stomata and continue to grow within the host tissue for 8–12 days without visible symptoms on the leaf surface (Goodwin et al., 2011). This biotrophic growth is followed by a switch to the necrotrophic growth phase which is characterized by a rapid increase in fungal biomass and emergence of STB symptoms including necrosis, chlorosis and typical brown black pycnidial lesions. To develop new disease control measures, it is crucial to understand the molecular mechanisms that underpin this developmental programme (Orton et al., 2011; Goodwin et al., 2011).
In Z. tritici, targeted gene deletion is the main strategy for functional genetic analysis and has been used to characterize various gene families, including members of those encoding secreted effector proteins (Motteram et al., 2009), mitogen activated protein kinases (Mehrabi et al., 2006a,b) and ATP-binding cassettes transporters (Zwiers et al., 2003). Agrobacterium tumefaciens mediated transformation (ATMT) is the most widely used method of introducing foreign DNA into the Z. tritici genome (Zwiers and De Waard, 2001; Bowler et al., 2010). As is the case for many ascomycete fungi such as Neurospora crassa (Ninomiya et al., 2004), Stagonospora nodorum (Feng et al., 2012) and Magnaporthe oryzae (Kershaw and Talbot, 2009) the gene targeting efficiency in Z. tritici is often low and locus dependent (Bowler et al., 2010) and is thus not amenable to a medium or high-throughput functional genomics analysis. Several physical factors such as chromosomal position, chromatin structure and transcriptional state of the target region affects the homologous recombination (HR) mediated integration of transforming DNA (Ninomiya et al., 2004; Feng et al., 2012). The non-homologous end joining (NHEJ) pathway of double strand DNA break repair also has a major impact on the frequency of HR (Walker et al., 2001). In most ascomycete fungi, disruption of the NHEJ pathway of DNA repair leads to elevated gene targeting efficiency because the HR pathway remains the only functional DNA break repair mechanism resulting in increased frequency of HR between the genome and foreign DNA (Ninomiya et al., 2004; Kershaw and Talbot, 2009). The eukaryotic NHEJ pathway is regulated by a protein complex comprising of the Ku70–Ku80 protein heterodimer and a DNA-dependent protein kinase catalytic subunit (Walker et al., 2001). Thus inactivation of either or both the Ku protein encoding genes leads to the disruption of the NHEJ pathway (Ninomiya et al., 2004).
In addition to low HR, the paucity of available positive selection markers is a significant constraint on large scale reverse genetics in Z. tritici. Three selectable marker genes, namely the phosphinothricin acetyltransferase encoding bar gene isolated from Streptomyces hygroscopicus (Thompson et al., 1987), the neomycin phosphotransferase II encoding nptII gene and the hygromycin phosphotransferase encoding hph gene (Gritz and Davies, 1983), both isolated from Escherichia coli, which confer resistance against glufosinate ammonium, geneticin and hygromycin B respectively, are routinely used in genetic manipulation of Z. tritici (Payne et al., 1998; Bowler et al., 2010; Zwiers and De Waard, 2001; Kramer et al., 2009). In addition, the carbendazim and carboxin resistant alleles of the β-tubulin encoding TUB1 and succinate dehydrogenase subunit B encoding MgSDHB gene from Z. tritici, respectively were also established as selectable markers (Payne et al., 1998; Bowler et al., 2010). However these markers are rarely used for genetic manipulations of Z. tritici. The mutation leading to carboxin resistance in Z. tritici (Scalliet et al., 2012) and carbendazim resistance in Fusarium graminearum (Zhang et al., 2009) are also known to affect in planta virulence in these organisms. This implies that the host derived marker genes are unsuitable for use in genetic manipulation to investigate traits associated with fungal pathogenicity and virulence. Additionally the geneticin resistance marker was used in a previous study to inactivate the Z. tritici KU70 gene (Bowler et al., 2010); therefore, only two markers conferring resistance to hygromycin and glufosinate ammonium remain for use in genetic manipulations of this Z. tritici ku70 null strain. Thus it is difficult to construct triple gene deletion mutants in this background. Such multiple gene deletion strains would be useful for characterizing complex traits associated with fungal infection, as, for example, in the analysis of secreted proteinases in Candida albicans and zinc transporters in Aspergillus fumigatus (Amich et al., 2010; Watts et al., 1998). Auxotrophic markers are also used in genetic manipulation of fungi. However, disruption of amino acid synthesis can affect fungal pathogenesis and therefore the use of auxotrophic markers should be avoided (de Gontijo et al., 2014; Sweigard et al., 1998). By increasing the number of selectable markers available for Z. tritici, it should be possible to facilitate the analysis of complex virulence phenotypes.
This report describes the construction and utility of Z. tritici ku70 and ku80 null strains in improving gene-targeting efficiency. The establishment of a sulfonylurea resistance cassette (ILV2SUR (GenBank accession AF013601), based on an allele of the Magnaporthe oryzae ILV2 gene, which encodes a variant of the acetolactate synthase enzyme that confers resistance to chlorimuron ethyl (a sulfonylurea herbicide), as a new dominant selectable marker system for Z. tritici (Valent and Chumley, 1991). In addition we have adapted the A. tumefaciens and E. coli binary vector backbone pCAMBIA-0380 to facilitate yeast recombinational cloning in Saccharomyces cerevisiae (Collopy et al., 2010; Gibson et al., 2008). Taken together these tools add significantly to our ability to genetically manipulate Z. tritici.
2. Materials and methods
2.1. Strains and growth conditions
The Z. tritici wild-type strain IPO323 was purchased from the Fungal Biodiversity Centre, Netherlands (http://www.cbs.knaw.nl). The IPO323 genome sequence was accessed at http://genome.jgi-psf.org/Mycgr3/Mycgr3.home.html (Goodwin et al., 2011). All reagents and chemicals were purchased from Sigma–Aldrich (Dorset, UK), unless otherwise stated.
All Z. tritici strains used in this study were cultured on Yeast Peptone Dextrose agar (YPD agar, per litre: 10 g yeast extract, 20 g peptone, 20 g dextrose and 20 g agar) or Basal medium (BM per litre: 1.7 g yeast nitrogen base without amino acids or ammonium sulphate (Formedium™, Hunstanton, UK), 2 g asparagine, 1 g NH4NO3, 10 g glucose and 20 g agar, pH to 6.0 with 1 M Na2HPO4) (Yang and Naqvi, 2014). The selection agent chlorimuron ethyl referred to as sulfonylurea hereafter (a gift from DuPont, Wilmington, USA) was dissolved in dimethylformamide to make a 2 mg/mL stock solution. Induction Medium agar (IM agar: 10 mM K2HPO4, 10 mM KH2PO4, 2.5 mM NaCl, 2 mM MgSO4.7H20, 0.7 mM CaCl2, 10 μM FeSO4, 4 mM (NH4)2SO4, 10 mM Glucose, 40 mM 2-N-morpholoino ethanesulfonilic acid, 0.5% Glycerol (w/v), 15 g agar/litre (w/v); pH to 5.6 with 1 M NaOH) supplemented with 200 μM acetosyringone was used for ATMT. For the mutagen sensitivity screen, Z. tritici strains were inoculated in synthetic complete (SC) broth (SC broth per litre: 6.9 g yeast nitrogen base without amino acids (Formedium™), 790 mg complete amino acid supplement mixture (Formedium™) and 20 g glucose) and cultures were grown at 18 °C and 200 RPM for 6 days. For microscopy, cultures were grown in SC, Czapek Dox broth (CDB) (Sigma–Aldrich, UK) or Aspergillus minimal media (AMM) broth (Bowler et al., 2010) for 72 h at 18 °C and 120 RPM. For ATMT and the sulfonylurea sensitivity screen, Z. tritici yeast like budding cells were grown on YPD agar plates incubated at 18 °C for 6 days, then harvested by adding sterile water and gently scraping the agar surface. Harvested cell suspensions were enumerated using a haemocytometer and suspensions were adjusted to required concentrations. S. cerevisiae strain BY4741 (MATa, his3, leu2, met15, ura3) was used for plasmid construction via recombinational cloning. A 50 ml YPD culture was inoculated with a single colony and grown at 30 °C and 180 RPM for 12 h to early stationary phase. Cells were harvested and used for standard transformations protocols (Gietz and Woods, 2002). The transformed cells were spread on SC dropout agar lacking uracil (SC-URA agar per litre: 6.9 g yeast nitrogen base with ammonium sulphate without amino acids (Formedium™), 790 mg complete amino acid supplement without uracil (Formedium™), 20 g glucose and 20 g agar) to select uracil prototrophs. Individual prototrophic colonies were identified after 48–72 h incubation at 30 °C. E. coli strain DH5α was used to propagate all plasmids. E. coli cultures were grown at 37 °C in Luria–Bertani (LB) media supplemented with 100 μg/mL kanamycin. A. tumefaciens strain EHA105 was used for the ATMT of Z. tritici. A. tumefaciens cultures were grown at 30 °C in LB or IM broth containing 100 μg/mL kanamycin and 100 μg/mL rifampicin.
2.2. Nucleic acid manipulations
Molecular cloning and DNA manipulations were carried out by following standard protocols (Sambrook and Russell, 2001), unless otherwise stated. All PCR primers are shown in Supplementary Table S1. Phusion® high fidelity polymerase (New England Biolabs (NEB), Herts, UK) was used to PCR amplify fragments for cloning while GoTaq® flexi DNA polymerase (Promega, Southampton, UK) was used for diagnostic colony PCR. All restriction endonucleases and T4-DNA ligase were purchased from NEB. Recombinant vectors were sequenced using the Sanger sequencing service at Eurofins Genomics (Ebersberg, Germany). Z. tritici genomic DNA was extracted in a Fastprep® cell disruptor (Fisher Scientific, Loughborough, UK) using a phenol chloroform extraction protocol (Motteram et al., 2009). Plasmids were extracted from E. coli using the QIAprep spin mini-prep kit (Qiagen, Manchester, UK). Recombinant plasmids were isolated from S. cerevisiae as described by Singh and Weil (2002).
2.3. Vector construction
The A. tumefaciens binary vector pC-HYG (Motteram et al., 2009) was modified by addition of the S. cerevisiae 2μ origin of replication and the URA3 selection marker to construct ternary vector pC-HYG-YR (Addgene ID – 61765) (see supplementary materials and methods). The ternary vectors pC-SUR-YR (Addgene ID – 61768), pC-BAR-YR (Addgene ID – 61766) and pC-G418-YR (Addgene ID – 61767) in which the hygromycin resistance marker of pC-HYG-YR was replaced with the markers which confer resistance against sulfonylurea, glufosinate ammonium and geneticin, respectively were constructed by yeast recombinational cloning (see supplementary materials and methods). To generate cassettes for inactivation of the KU70 and KU80 genes, left flanks (LF) and right flanks (RF) of each gene were cloned by yeast recombinational cloning on either side of the resistance marker in the vectors pC-G418-YR and pC-SUR-YR to construct pC-G418-KU70-KO, pC-G418-KU80-KO and pC-SUR-KU80-KO (see supplementary materials and methods). Similarly, vectors pC-HYG-36951-KO, pC-HYG-72646-KO and pC-HYG-102083-KO used to inactivate the Z. tritici genes Mycgr3G36951 (encoding a putative non-ribosomal peptide synthetase), Mycgr3G72646 (encoding a putative α-(1,3)-glucan synthase) and Mycgr3G102083 (encoding putative isocitrate lyase) respectively were constructed using vector pC-HYG-YR and yeast recombinational cloning (see supplementary materials and methods).
2.4. A. tumefaciens mediated transformation of Z. tritici
The vectors pC-G418-KU70-KO, pC-G418-KU80-KO, pC-SUR-KU80-KO, pC-HYG-36951-KO, pC-HYG-72646-KO, pC-HYG-102083-KO and pC-SUR-YR were introduced into chemically competent cells of A. tumefaciens strain EHA105 (Flowers and Vaillancourt, 2005). ATMT of Z. tritici was carried out with minor modifications to standard protocols (Zwiers and De Waard, 2001; Motteram et al., 2009). Briefly, A. tumefaciens cultures (OD595 = 0.25–0.30) carrying the appropriate vector and a Z. tritici cell suspension (1 × 107 cells/mL in sterile water) were mixed in a ratio of 3:1 and 400 μL of this mixture was spread on nitrocellulose membranes (A.A. Packaging Limited, Preston, UK) placed on IM agar. After 48 h incubation at 25 °C, the nitrocellulose discs were transferred, as appropriate for each vector, onto selection plates containing BM agar amended with 10 μg/mL sulfonylurea or YPD agar containing 200 μg/mL geneticin (G418 sodium salt) or hygromycin B and antibiotics (100 μg/mL ampicillin, 100 μg/mL cefotaxime, 100 μg/mL streptomycin and 100 μg/mL timentin). After 14 days putative drug resistant Z. tritici transformants were sub cultured for two rounds of selection on BM agar containing 10 μg/mL sulfonylurea or YPD agar containing 200 μg/mL geneticin or hygromycin and confirmed drug resistant strains were selected for further analysis.
To confirm the functionality of the sulfonylurea resistance cassette, fifteen independent sulfonylurea resistant transformants harbouring the ternary vector pC-SUR-YR were isolated and designated as strains IPO323:SurR 1–15. The genomic integration of the sulfonylurea resistance cassette in strains IPO323:SurR 1–15 was confirmed by diagnostic PCR using primers pSURR-FWD and SURR-INT-R which amplify a single 558 bp fragment specific to the resistance cassette.
Putative Z. tritici ku70 and ku80 null strains resistant to geneticin and harbouring pC-G418-KU70-KO and pC-G418-KU80-KO were isolated. These strains were designated HLS1000 (Δku70:G418R) and HLS1001 (Δku80:G418R) respectively. Similarly to demonstrate the application of sulfonylurea resistance cassette as a marker for targeted gene deletion in Z. tritici, sulfonylurea resistant strains transformed with pC-SUR-KU80-KO were isolated and designated HLS1002 (Δku80:SurR). To confirm the inactivation of the Z. tritici KU70 and KU80 genes, drug resistant transformants were analysed by diagnostic PCR using primer combinations KU70-EXT-F/KU70-INT-R/G418R-INT-R, KU80-EXT-F/KU80-INT-R/G418R-INT-R or KU80-EXT-F/KU80-INT-R/SURR-INT-R. To confirm the integration events of deletion vectors into strains HLS1000, HLS1001 and HLS1002, Southern blot analysis was performed as described by Motteram et al. (2009), Sambrook and Russell (2001). Briefly genomic DNA isolated from Z. tritici strains HLS1000, HLS1001 and IPO323 was digested with PvuI and HindIII followed by transfer onto nylon membranes. A 152 bp long DIG-dUTP labelled DNA probe was synthesized using PCR primers Probe-F/R which are specific to the geneticin resistance cassette (Supplementary Table S1) following the manufacturers’ instruction using PCR DIG probe synthesis kit (Roche Diagnostics Ltd., Burgess Hill, UK). After incubation of the membrane in the probe hybridization solution for 12 h at 65 °C, the membrane was washed as described by Motteram et al. (2009) and images were acquired using a GBOX Chemi XX6 imaging system (Syngene, Cambridge, UK). A single hybridization product at 2409 bp and 2887 bp confirmed the deletion of the KU70 and KU80 gene respectively without any events of ectopic integration of the deletion construct. Similarly, another Southern blot analysis was also performed on the genomic DNA isolated from strains HLS1002 and IPO323 which was digested with restriction endonucleases BstEII and SphI. The DIG-dUTP labelled probe specific to the KU80 right flank was synthesized using primers KU80-Probe-F/R (Supplementary Table S1). A single hybridization product at 3792 bp indicated deletion of the KU80 gene in strain HLS1002 while a single hybridization band at 7241 bp indicated the wild type KU80 gene.
To compare gene targeting efficiency in strains HLS1000 and HLS1001 to the wild type strain IPO323, hygromycin resistant strains from each strain background transformed with pC-HYG-36951-KO, pC-HYG-72646-KO and pC-HYG-102083-KO were isolated. Diagnostic PCR was carried out using the primer pair 36951-EXT-F/36951-INT-R which amplified a 1735 bp product specific to the Mycrg3G36951 wild type allele or a 2890 bp product amplified using 36951-EXT-F/HYGR-INT-R indicating replacement of the Mycrg3G36951 gene by the hygromycin resistance cassette. Similarly primer combination 72646-EXT-F/72646-INT-R and 102083-EXT-F/102083-INT-R amplified 1200 and 1400 bp products from wild type alleles of Mycgr3G72646 and Mycgr3G102083 respectively. The amplification of 1900 bp and 2100 bp PCR products using the primer pairs 72646-EXT-F/HYGR-INT-R and 102083-EXT-F/HYGR-INT-R confirmed the replacement of Mycgr3G72646 and Mycgr3G102083 by the hygromycin resistance cassette.
2.5. Sulfonylurea sensitivity screen
To titrate the optimal inhibitory concentration for selection with sulfonylurea, the Z. tritici wild type strain IPO323 and strain IPO323:SurR 1 were sub-cultured 10 times on non-selective YPD agar and then used for drug sensitivity screens. For each strain, yeast like cells were harvested and a 10 μL aliquot of cell suspension (serially diluted to 106, 105 and 104 cells/mL in sterile water) was spotted in triplicate on BM agar containing sulfonylurea concentrations ranging from 0 to 12 μg/mL. Plates were incubated at 18 °C for 6 days and images were acquired using a GBOX Chemi XX6 imaging system (Syngene).
2.6. Phenotypic characterization of the Z. tritici ku70 and ku80 null strains
Z. tritici strains HLS1000, HLS1001, HLS1002 and IPO323 were screened to identify phenotypes that may arise from the deletion of KU70 or KU80 genes. Mutagen sensitivity screens were carried out essentially as described by Ninomiya et al. (2004). Cells were spotted on SC agar amended with mutagens and plates were incubated at 18 °C for 6 days and images were acquired using a GBOX Chemi XX6 imaging system (Syngene, UK). For microscopic analysis, Z. tritici cells were harvested from cultures grown in SC, CDB and AMM and images were acquired using an Olympus IX8I spinning disc microscope (Olympus, Southend-on-Sea, UK). To determine the impact of Z. tritici KU70 and KU80 gene deletion on pathogenicity, whole plant infection assays (Motteram et al., 2009) were carried out. Briefly, the second leaf of 14 day-old wheat plants (susceptible cultivar Avalon) was inoculated with yeast like budding cells (1 × 107 cells/ml in water containing 0.1% (v/v) Tween 20) of Z. tritici strains HLS1000, HLS1001, HLS1002 and the wild type strain IPO323. Mock infection was carried out using sterile water containing 0.1% (v/v) Tween20. Infected plants were maintained at 16:8 h day:night cycles at 18 °C and 85% relative humidity for 21 days. Visual inspections of infection were carried out at 14, 18 and 21 days after infection (DAI). Images were acquired with a Leica M205FA stereo microscope (Leica, Milton Keynes, UK) and Epson scanner (Epson, Hertfordshire, UK).
3. Results and discussion
3.1. Yeast recombinational cloning for vector construction
In a recent study, we have utilized yeast recombinational cloning to construct thirty two Z. tritici over-expression vectors (Sidhu et al., 2015). This technique circumvents resource-heavy conventional cloning which is often used to construct gene deletion vectors for Z. tritici. We therefore developed four A. tumefaciens ternary vectors for recombinational cloning in S. cerevisiae. The A. tumefaciens binary vector pC-HYG (Motteram et al., 2009) was modified by adding the S. cerevisiae 2μ origin of replication and the URA3 selection marker to give pC-HYG-YR (Fig. S1). The hygromycin resistance cassette in pC-HYG-YR was replaced with the ILV2SUR sulfonylurea resistance cassette from M. oryzae (Valent and Chumley, 1991), the bar gene for resistance against glufosinate ammonium (Kramer et al., 2009) or the nptII gene for resistance against geneticin (Bowler et al., 2010). The resulting sequence-verified vectors were designated pC-SUR-YR, pC-BAR-YR and pC-G418-YR, respectively (Fig. S1).
3.2. Sensitivity to sulfonylurea and functionality of the resistance cassette in Z. tritici
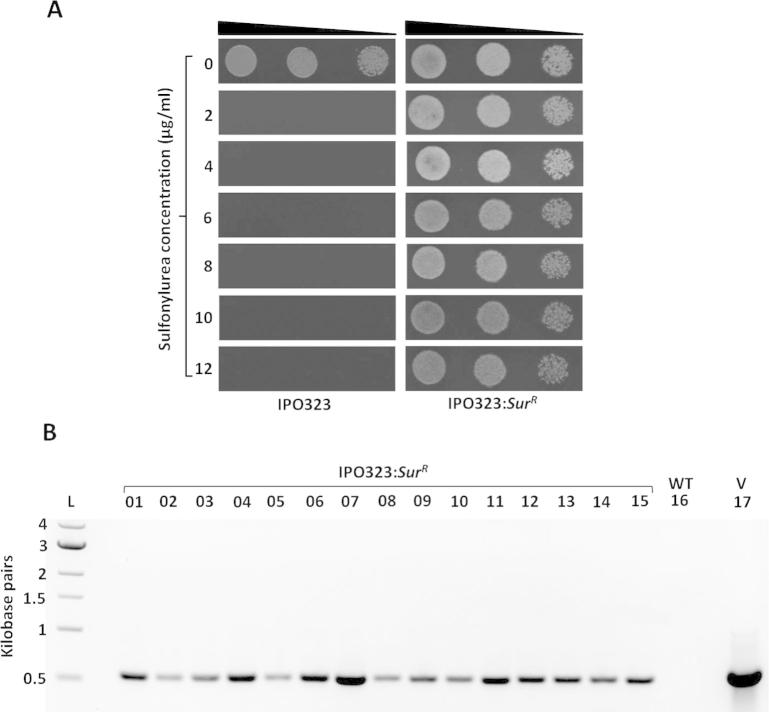
To establish the sulfonylurea resistance cassette as a new positive selection marker for use in Z. tritici it was necessary to confirm its sensitivity to sulfonylurea. Z. tritici wild type strain IPO323 was highly sensitive to sulfonylurea and growth was completely inhibited at drug concentrations as low as 2 μg/mL (Fig. 1). However, for selection on cellophane discs, which are used for ATMT transformation of Z. tritici, we observed that 10 μg/mL sulfonylurea was needed to eliminate background Z. tritici IPO323 growth (data not shown).
Fig. 1.
Z. tritici wild type strain IPO323 is hypersensitive to sulfonylurea (A) and genomically integrated ILV2SUR sulfonylurea resistance cassette confers resistance to the drug (B). (A) Z. tritici wild type strain IPO323 is hypersensitive to sulfonylurea (chlorimuron ethyl is the active ingredient) at low concentrations (2 μg/mL) while strains IPO323:SurR (independent mutants containing a randomly integrated sulfonylurea resistance cassette) are drug resistant following 10 rounds of growth on non-selective media. Z. tritici cells (from 106 to 104 cells/ml) were spotted on BM agar supplemented with various sulfonylurea concentrations and plates were incubated at 18 °C for 6 days. The wild type strain IPO323 shows severe growth inhibition whereas the IPO323:SurR mutant is resistant to sulfonylurea concentrations up to 12 μg/ml. (B) Agarose gel (1% w/v in TAE) image showing PCR confirmation of presence of the sulfonylurea resistance cassette in the genomic DNA isolated from 15 randomly selected IPO323:SurR mutants (Lane 1–15). Amplification of a 558 bp PCR product confirms that the sulfonylurea resistance cassette is present in strains IPO323:SurR 1–15, but absent in wild-type Z. tritici IPO323 (Lane 16/WT). The pC-SUR-YR vector Lane 17/V was used as positive control for amplification of the 558 bp product from the sulfonylurea resistance cassette.
To confirm the functionality of the sulfonylurea resistance cassette in Z. tritici, the vector pC-SUR-YR was transformed into strain IPO323 and 15 sulfonylurea resistant transformants designated strain IPO323:SurR 1–15 were isolated after 14 days growth on BM agar. Diagnostic PCR was carried out on the genomic DNA to confirm the genomic integration of the resistance cassette and a 558 bp fragment specific to the sulfonylurea resistance cassette was amplified from strains IPO323:SurR 1–15 but the PCR product was absent in case of wild type Z. tritici IPO323 (Fig. 1). Further, a single IPO323:SurR strain was sub-cultured for 10 rounds on non-selective medium and then spotted on BM agar containing 1–12 μg/ml sulfonylurea (Fig. 1). This strain retained drug resistance suggesting genomic integration of the sulfonylurea resistance cassette was mitotically stable even in absence of selection pressure. These data demonstrate that the sulfonylurea resistance cassette from M. oryzae is a functional dominant selection marker in Z. tritici.
3.3. Inactivation of the Z. tritici KU70 and KU80 genes
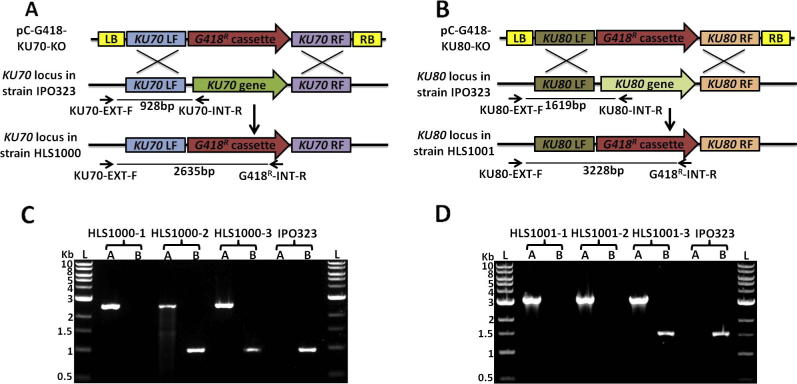
To inactivate the predicted Z. tritici KU70 (Gene ID Mycgr3G85040) and KU80 (Gene ID Mycgr3G40048) genes, the respective gene deletion vectors pC-G418-KU70-KO, pC-G418-KU80-KO and pC-SUR-KU80-KO were introduced into the wild type strain IPO323 via ATMT. Diagnostic PCR was used to test for replacement of KU70 by the geneticin resistance cassette (Fig. 2A), using the primer pairs KU70-EXT-F/KU70-INT-R and KU70-EXT-F/G418R-INT-REV. Out of 96 putative transformants tested, the amplification of a 2635 bp product in three suggested the replacement of KU70 by the geneticin resistance cassette (Fig. 2C). However, amplification of a 928 bp product, using KU70-EXT-F/KU70-INT-R, in two out of these three putative ku70 null strains also suggested the presence of a wild type KU70 allele (Fig. 2C). The deletion of the KU80 gene was confirmed by diagnostic PCR on another set of 96 putative mutants, harbouring the vector pC-G418-KU80-KO (Fig. 2B). A 3228 bp PCR product amplified using primers KU80-EXT-F/G418R-INT-REV from the genomic DNA isolated from three mutants suggested the KU80 gene had been replaced by the geneticin resistance cassette (Fig. 2D). However, amplification of a 1619 bp PCR product using primers KU80-EXT-F/KU80-INT-REV in one of these putative ku80 null mutants suggested the presence of wild type KU80 gene (Fig. 2D). Southern blot analysis confirmed that in the ku70 and ku80 null strains the target genes had been replaced by the geneticin resistance cassette without any ectopic integrations events (Fig. S2). These confirmed ku70 and ku80 null strains were designated Z. tritici HLS1000 and HLS1001, respectively.
Fig. 2.
Construction of Z. tritici ku70 and ku80 null strains. Z. tritici KU70 and KU80 gene deletion vectors pC-G418-KU70-KO and pC-G418-KU80-KO were introduced into the wild type strain IPO323 by A. tumefaciens mediated transformation. Schematic representations of the gene deletion strategies for Z. tritici KU70 (A) and KU80 (B) are shown. A. tumefaciens left (LB) and right (RB) border regions (yellow) on the transfer DNA (TDNA) flank the deletion constructs. Homologous recombination between sequences in the left flank (KU70LF or KU80LF) and right flank (KU70RF or KU80RF) of the deletion cassette and the chromosomal KU loci mediate integration into their respective target locus to replace KU70 and KU80 with the geneticin resistance marker (G418R cassette) resulting in ku70 and ku80 null strains HLS1000 and HLS1001, respectively. (C and D) Image of ethidium bromide stained agarose gel (2% in TAE (w/v)) shows the results of the diagnostic PCR on putative ku70 and ku80 null strains. Primer pair KU70-EXT-F/KU70-INT-R and KU70-EXT-F/G418R-INT-R amplify 928 bp and 2635 bp products which confirm the presence of KU70 wild type gene and replacement of the KU70 by the geneticin resistance marker, respectively. Similarly, primer pairs KU80-EXT-F/KU80-INT-R and KU80-EXT-F/G418R-INT-R amplify a 1619 bp product specific to the KU80 wild type gene and amplification of a 3228 bp product indicates replacement of the KU80 gene by the geneticin resistance marker. The NEB 1 kb DNA ladder (L) was used as a standard. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We also attempted to inactivate Z. tritici KU80 with pC-SUR-KU80-KO to test the utility of the sulfonylurea resistance cassette as a positive selection marker for gene deletion. The vector pC-SUR-KU80-KO was transformed into the wild type strain Z. tritici IPO323. Colony PCR was carried out, using a combination of three primers KU80-EXT-F/KU80-INT-R/SURR-INT-R (see Supplementary Table S1) to identify ku80 null strains (Fig. S3A). Out of 192 transformants that were screened, four showed a single 2490 bp amplicon, which confirmed that the deletion construct had integrated into the KU80 locus (Fig. S3B). Southern blot analysis showed that the DIG-dUTP labelled probe hybridized at a single 3792 bp band thus confirming gene deletion without ectopic integration of the deletion vector (Fig. S3C). The ku80 null strain constructed using the sulfonylurea resistance cassette was designated HLS1002. These experiments confirmed that the sulfonylurea resistance cassette can be used as a positive selection marker for gene deletion in Z. tritici.
3.4. Characterization of the Z. tritici ku70 and ku80 null strains
To test whether inactivation of the Z. tritici KU70 and KU80 gene in strains HLS1000 and HLS1001 respectively affected gene targeting frequency, we attempted to inactivate three genes (Table 1). Using ATMT each gene deletion construct was individually introduced into strains HLS1000, HLS1001 and wild type Z. tritici IPO323. Diagnostic PCR was carried out on 16 hygromycin resistant transformants isolated from each background (HLS1000, HLS1001 and IPO323). The deletion frequency of the 13,961 bp long gene Mycgr3G36951 exceeded 90% in strains HLS1000 and HLS1001 as compared to 6.25% observed in the IPO323 background (Fig. S4 and Table 1). Similarly, the other two genes Mycgr3G72646 and Mycgr3G102083 were also targeted at an elevated frequency exceeding 85% in strains HLS1000 and HLS1001 as compared to less than 10% in the wild type strain (Table 1). These results reveal a dramatic increase in the gene targeting frequency as a result of inactivation of both the Z. tritici KU70 and KU80 genes. This is in agreement with previously reported increased gene deletion frequencies that were achieved in an independently created Z. tritici ku70 null strain (Bowler et al., 2010). In addition, our ku70 null strain HLS1000 was also successfully used for high frequency targeted integration of gene overexpression constructs (Sidhu et al., 2015) and to replace the native promoter of the Z. tritici β-1,3-glucan synthase gene (Marchegiani et al., 2015).
Table 1.
Gene targeting frequency in Z. tritici strains HLS1000 (ku70 null), HLS1001 (ku80 null) and IPO323 (wild type). (Frequency was measured as percentage of integrations into target locus).
| Gene name/ID | Size (bp) | Locus | HLS1000 (%) | HLS1001 (%) | IPO323 (%) |
|---|---|---|---|---|---|
| Mycgr3G36951 | 13,961 | chr_2:439283-453243 | 93.75 | 100 | 6.25 |
| Mycgr3G72646 | 7,932 | chr_5:2631468-2639399 | 100 | 87.50 | 0 |
| Mycgr3G102083 | 2,058 | chr_1:1020739-1022796 | 100 | 93.75 | 6.25 |
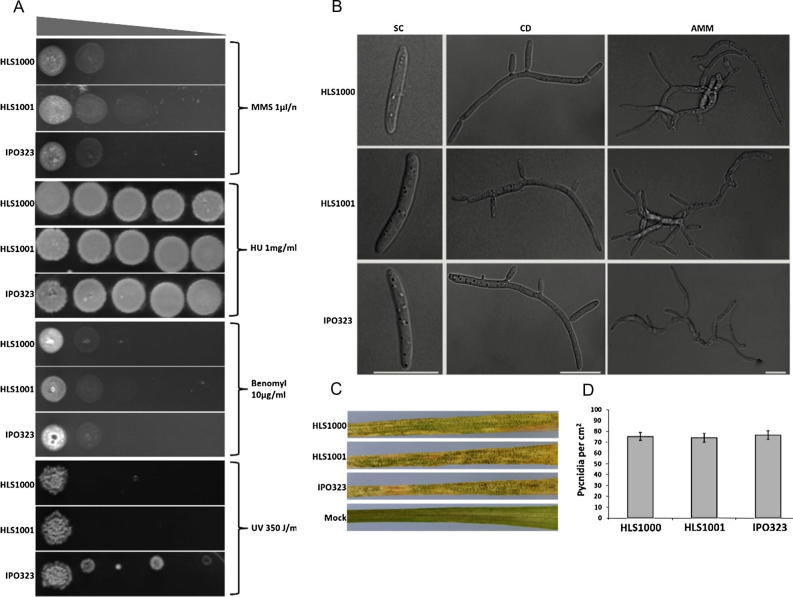
Due to disruption of DNA repair, the NHEJ mutants can become hypersensitive to DNA damaging mutagens (Hoff et al., 2010). In line with observations in N. crassa (Ninomiya et al., 2004), the Z. tritici strains HLS1000 and HLS1001 showed mildly increased sensitivity to ultraviolet (UV) radiation but not to methyl methane sulphate (MMS), hydroxyurea (HU) or benomyl (Fig. 3A). These compounds result in point mutations but not in double strand breaks in DNA and therefore it is not surprising that both mutants are not hypersensitive to these mutagens. Strains HLS1000 and HLS1001 showed no obvious morphological growth defects in standard in vitro conditions (Fig. 3B). Similar to wild type Z. tritici IPO323, both strains exhibited yeast like cell growth in SC medium while pseudohyphal and hyphal growth was predominant when strains were grown in nutrient limiting CDM and AMM (Fig. 3B).
Fig. 3.
Z. tritici ku70 and ku80 null mutants are essentially wild type in respect of mutagen sensitivity, in vitro growth and pathogenicity. (A) Z. tritici strains HLS1000 (ku70 null) and HLS1001 (ku80 null) are essentially wild-type in their response to methyl methanesulfonate (MMS), benomyl and hydroxyurea (HU), but exhibit mildly increased sensitivity to ultraviolet (UV) radiation as compared to the wild type strain IPO323. Serially diluted cells (108–104 cells/mL) were spotted on SC agar containing MMS (1 μl/mL), HU (1 mg/ml), benomyl (10 μg/mL) or exposed to UV radiation (350 J/m2). (B) Deletion of the Z. tritici KU70 and KU80 genes does not affect in vitro growth. Strains HLS1000, HLS1001 and IPO323 were grown in SC, CD or AMM for 72 h at 18 °C and 120 RPM. Images were acquired in the bright field channel using an Olympus IX8I spinning disc microscope and Visiview imaging suite. Scale bar represents 10 μm. (C) Plant infection assay shows that strains HLS1000 and HLS1001 are pathogenic and induce typical symptoms (necrosis, chloresis and formation of black/brown pycnidial lesions) of Z. tritici infection similar to the wild type strain IPO323. Whole leaves of wheat plants of susceptible cultivar Avalon were inoculated with 1 × 107 cells/mL and images were acquired 21 days after infection. (D) Quantitative analysis of infection (pycnidia per cm2) showed no significant difference in pathogenicity of strain HLS1000 (p = 0.25) and HLS1001 (p = 0.55) as compared to wild type strain IPO323. Pycnidia were counted manually and data was statistically analysed using analysis of variance (ANOVA).
To be useful for in vivo analysis it is imperative that mutations that facilitate homologous recombination (such as deletion of KU70 and KU80) and selectable markers (such as geneticin and sulfonylurea resistance cassettes) used for transformation of pathogenic fungi, do not detectably impact virulence during infection assays. To determine this for these strains wheat leaves (susceptible cultivar Avalon) were inoculated with Z. tritici HLS1000, HLS1001, HLS1002 and the wild type progenitor strain IPO323. Visual inspection of infection revealed no differences in the initiation of chlorosis at 14 DAI, in appearance of typical black brown pycnidial lesions at 18 DAI and necrosis at 21 DAI on the leaves infected with any strain (Figs. 3C and S3D). Quantification of infection suggested that inactivation of KU70 or KU80 did not affect virulence and both Z. tritici HLS1000 and HLS1001 were as pathogenic as the wild type strain IPO323 (Fig. 3D). Microscopic observation at 21 DAI revealed secretion of pycnidiospore-bearing cirrhi from the characteristic black-brown pycnidial lesions formed on leaves infected by strains HLS1002 and IPO323 (Fig. S3E, data not shown for strains HLS1000 and HLS1001). The similar progression of infection and specifically the presence of pycnidiospores, the asexual spores required for initiation of multiple STB infection cycles during the growth season (Orton et al., 2011) demonstrate that neither expression of the resistance cassette, nor the deletion of the Z. tritici KU70 or KU80 genes detectably affect pathogenicity in planta.
The Z. tritici KU70 and KU80 genes were deleted to increase the HR frequency and improve gene targeting. This objective was successfully accomplished as both Z. tritici HLS1000 and HLS1001 facilitated targeted gene inactivation at higher efficiency (on average >90%) than wild type, for the three different genomic loci that were tested. These results also confirm that NHEJ is the dominant pathway of double stranded DNA break repair in Z. tritici. This strengthens the proposition that in contrast to S. cerevisiae where DNA damage is predominantly repaired by HR (Rothstein, 1991) in most filamentous fungi the NHEJ pathway mediates DNA damage repair (Villalba et al., 2008; Ninomiya et al., 2004). The phenotypic screen confirmed that the in vitro growth and in planta pathogenicity of the ku70 and ku80 null strains is comparable to the wild type strain. These results suggest that the ku70 and ku80 null strains are an ideal platform for construction of gene deletion mutants in Z. tritici. In N. crassa and M. oryzae the NHEJ mutants have proved to be an excellent platform to dissect novel roles of multiple transcription factors (Colot et al., 2006) and authophagy (Kershaw and Talbot, 2009). N. crassa NHEJ mutants also enabled construction of the whole genome deletion library (Collopy et al., 2010) which is an invaluable resource for providing new insights into molecular and cell biology (Fu et al., 2011; Berepiki and Read, 2013). Thus, the Z. tritici ku70 and ku80 null strains and other tools reported in this study should play an important role in furthering functional analysis in this important pathogen.
4. Conclusion
In this study, we report the generation of new tools for construction of Z. tritici mutant strains. The confirmation of the functionality of the sulfonylurea resistance marker in Z. tritici increases the number of positive selection markers available for use in this fungus. Along with the ternary vectors offering yeast recombinational cloning, this represents a significant addition to the toolkit available for genetic manipulation of Z. tritici. ATMT is widely used to transform fungi and plants (Michielse et al., 2005) and therefore these vectors will also be useful for the wider fungal research community. The Z. tritici ku70 and ku80 null strains facilitate increased HR mediated gene targeting while maintaining wild type growth and pathogenicity in planta. Taken together, these tools complement currently available techniques and are a major step towards making large-scale gene functional analysis feasible in Z. tritici.
Acknowledgments
We thank Sreedhar Kilaru for technical assistance with the yeast recombinational cloning. This work was funded by a BBSRC BBR Grant to KH, NT & DS (BB/I025956/1) and a BBSRC CASE studentship to KH (BB/J500793/1), supported by Syngenta UK.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.fgb.2015.04.015.
Appendix A. Supplementary material
References
- Amich J., Vicentefranqueira R., Leal F., Calera J.A. Aspergillus fumigatus survival in alkaline and extreme zinc-limiting environments relies on the induction of a zinc homeostasis system encoded by the zrfC and aspf2 genes. Eukaryot. Cell. 2010;9:424–437. doi: 10.1128/EC.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berepiki A., Read N.D. Septins are important for cell polarity, septation and asexual spore formation in Neurospora crassa and show different patterns of localisation at germ tube tips. PLoS One. 2013;8:e63843. doi: 10.1371/journal.pone.0063843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler J., Scott E., Tailor R., Scalliet G., Ray J., Csukai M. New capabilities for Mycosphaerella graminicola research. Mol. Plant Pathol. 2010;11:691–704. doi: 10.1111/j.1364-3703.2010.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collopy P., Colot H., Park G., Ringelberg C., Crew C., Borkovich K., Dunlap J. High-throughput construction of gene deletion cassettes for generation of Neurospora crassa knockout strains. In: Sharon A., editor. Humana Press; 2010. (Molecular and Cell Biology Methods for Fungi). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H.V., Park G., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gontijo F.A., Pascon R.C., Fernandes L., Machado J., Jr., Alspaugh J.A., Vallim M.A. The role of the de novo pyrimidine biosynthetic pathway in Cryptococcus neoformans high temperature growth and virulence. Fungal Genet. Biol. 2014;70:12–23. doi: 10.1016/j.fgb.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Li W.J., Hwang S.F., Gossen B.D., Strelkov S.E. Enhanced gene replacement frequency in KU70 disruption strain of Stagonospora nodorum. Microbiol. Res. 2012;167:173–178. doi: 10.1016/j.micres.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Flowers J.L., Vaillancourt L.J. Parameters affecting the efficiency of Agrobacterium tumefaciens-mediated transformation of Colletotrichum graminicola. Curr. Genet. 2005;48:380–388. doi: 10.1007/s00294-005-0034-1. [DOI] [PubMed] [Google Scholar]
- Fu C., Iyer P., Herkal A., Abdullah J., Stout A., Free S.J. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot. Cell. 2011;10:1100–1109. doi: 10.1128/EC.05003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Benders G.A., Axelrod K.C., Zaveri J., Algire M.A., Moodie M., Montague M.G., Venter J.C., Smith H.O., Hutchison C.A. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Natl. Acad. Sci. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Goodwin S.B., Ben M’barek S., Dhillon B., Wittenberg A.H.J., Crane C.F., Hane J.K., Foster A.J., Van der Lee T.A.J., Grimwood J., Aerts A., Antoniw J., Bailey A., Bluhm B., Bowler J., Bristow J., Van der Burgt A., Canto-Canche B., Churchill A.C.L., Conde-Ferraez L., Cools H.J., Coutinho P.M., Csukai M., Dehal P., De Wit P., Donzelli B., Van de Geest H.C., Van Ham R., Hammond-Kosack K.E., Henrissat B., Kilian A., Kobayashi A.K., Koopmann E., Kourmpetis Y., Kuzniar A., Lindquist E., Lombard V., Maliepaard C., Martins N., Mehrabi R., Nap J.P.H., Ponomarenko A., Rudd J.J., Salamov A., Schmutz J., Schouten H.J., Shapiro H., Stergiopoulos I., Torriani S.F.F., Tu H., De Vries R.P., Waalwijk C., Ware S.B., Wiebenga A., Zwiers L.H., Oliver R.P., Grigoriev I.V., Kema G.H.J. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 2011;7(6):e1002070. doi: 10.1371/journal.pgen.1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritz L., Davies J. Plasmid-encoded hygromycin B resistance. the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- Hoff B., Kamerewerd J., Sigl C., Zadra I., Kuck U. Homologous recombination in the antibiotic producer Penicillium chrysogenum: strain ΔPcku70 shows up-regulation of genes from the HOG pathway. Appl. Microbiol. Biotechnol. 2010;85:1081–1094. doi: 10.1007/s00253-009-2168-4. [DOI] [PubMed] [Google Scholar]
- Kershaw M.J., Talbot N.J. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. U.S.A. 2009;106:15967–15972. doi: 10.1073/pnas.0901477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., Thines E., Foster A.J. MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea. Fungal Genet. Biol. 2009;46:667–681. doi: 10.1016/j.fgb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Marchegiani, E., Sidhu, Y.S., Haynes, K., Lebrun, M.H., 2015. Conditional gene expression and promoter replacement in Zymoseptoria tritici using fungal nitrate reductase promoters. Fungal Genet. Biol. 79, 174–179. [DOI] [PubMed]
- Mehrabi R., van der Lee T., Waalwijk C., Kema G.H.J. MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol. Plant Microbe Interact. 2006;19:389–398. doi: 10.1094/MPMI-19-0389. [DOI] [PubMed] [Google Scholar]
- Mehrabi R., Zwiers L.-H., de Waard M.A., Kema G.H.J. MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 2006;19:1262–1269. doi: 10.1094/MPMI-19-1262. [DOI] [PubMed] [Google Scholar]
- Michielse C.B., Hooykaas P.J.J., van den Hondel C.A., Ram A.F.J. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 2005;48:1–17. doi: 10.1007/s00294-005-0578-0. [DOI] [PubMed] [Google Scholar]
- Motteram J., Kufner I., Deller S., Brunner F., Hammond-Kosack K.E., Nurnberger T., Rudd J.J. Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain-containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 2009;22:790–799. doi: 10.1094/MPMI-22-7-0790. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y., Suzuki K., Ishii C., Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orton E.S., Deller S., Brown J.K.M. Mycosphaerella graminicola: from genomics to disease control. Mol. Plant Pathol. 2011;12:413–424. doi: 10.1111/j.1364-3703.2010.00688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne A.C., Grosjean-Cournoyer M.-C., Hollomon D.W. Transformation of the phytopathogen Mycosphaerella graminicola to carbendazim and hygromycin B resistance. Curr. Genet. 1998;34:100–104. doi: 10.1007/s002940050372. [DOI] [PubMed] [Google Scholar]
- Polley R.W., Thomas M.R. Surveys of diseases of winter wheat in England and Wales 1976–88. Annals Appl. Biol. 1991;119:1–20. [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Scalliet G., Bowler J., Luksch T., Kirchhofer-Allan L., Steinhauer D., Ward K., Niklaus M., Verras A., Csukai M., Daina A., Fonné-Pfister R. Mutagenesis and functional studies with succinate dehydrogenase inhibitors in the wheat pathogen Mycosphaerella graminicola. PLoS One. 2012;7:e35429. doi: 10.1371/journal.pone.0035429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu, Y.S., Chaudhari, Y.K., Usher, J., Cairns, T.C., Talbot, N.J., Studholme, D.J., Csukai, M., Haynes, K., 2015. A suite of Gateway® compatible ternary expression vectors for functional analysis in Zymoseptoria tritici. Fungal Genet. Biol. 79, 180–185. [DOI] [PubMed]
- Singh M.V., Weil A.P. A method for plasmid purification directly from yeast. Anal. Biochem. 2002;307:13–17. doi: 10.1016/s0003-2697(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Sweigard J.A., Carroll A.M., Farrall L., Chumley F.G., Valent B. Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol. Plant Microbe Interact. 1998;11:404–412. doi: 10.1094/MPMI.1998.11.5.404. [DOI] [PubMed] [Google Scholar]
- Thompson C.J., Movva N.R. Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 1987;6:2519–2523. doi: 10.1002/j.1460-2075.1987.tb02538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent B., Chumley F.G. Molecular genetic analysis of the rice blast fungus Magnaporthe grisea. Ann. Rev. Phytopathol. 1991;29:443–467. doi: 10.1146/annurev.py.29.090191.002303. [DOI] [PubMed] [Google Scholar]
- Villalba F., Collemare J., Landraud P., Lambou K., Brozek V., Cirer B., Morin D., Bruel C., Beffa R., Lebrun M.H. Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet. Biol. 2008;45:68–75. doi: 10.1016/j.fgb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Walker J.R., Corpina R.A., Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Watts H.J., Cheah F.S., Hube B., Sanglard D., Gow N.A. Altered adherence in strains of Candida albicans harbouring null mutations in secreted aspartic proteinase genes. FEMS Microbiol. Lett. 1998;159:129–135. doi: 10.1111/j.1574-6968.1998.tb12851.x. [DOI] [PubMed] [Google Scholar]
- Yang F., Naqvi N.I. Sulfonylurea resistance reconstitution as a novel strategy for ILV2-specific integration in Magnaporthe oryzae. Fung. Genet. Biol. 2014;68:71–76. doi: 10.1016/j.fgb.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Zhang Y.J., Yu J.J., Zhang Y.N., Zhang X., Cheng C.J., Wang J.X., Hollomon D.W., Fan P.S., Zhou M.G. Effect of carbendazim resistance on trichothecene production and aggressiveness of Fusarium graminearum. Mol. Plant Microbe Interact. 2009;22:1143–1150. doi: 10.1094/MPMI-22-9-1143. [DOI] [PubMed] [Google Scholar]
- Zwiers L.-H., de Waard M. Efficient Agrobacterium tumefaciens-mediated gene disruption in the phytopathogen Mycosphaerella graminicola. Curr. Genet. 2001;39:388–393. doi: 10.1007/s002940100216. [DOI] [PubMed] [Google Scholar]
- Zwiers L.H., Stergiopoulos I., Gielkens M.M.C., Goodall S.D., de Waard M.A. ABC transporters of the wheat pathogen Mycosphaerella graminicola function as protectants against biotic and xenobiotic toxic compounds. Mol. Genet. Genomics. 2003;269:499–507. doi: 10.1007/s00438-003-0855-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.