Figure 1. The role of DNA-PKcs in the non-homologous end joining (NHEJ) pathway for the repair of ionizing radiation induced DNA double strand breaks (DSBs).
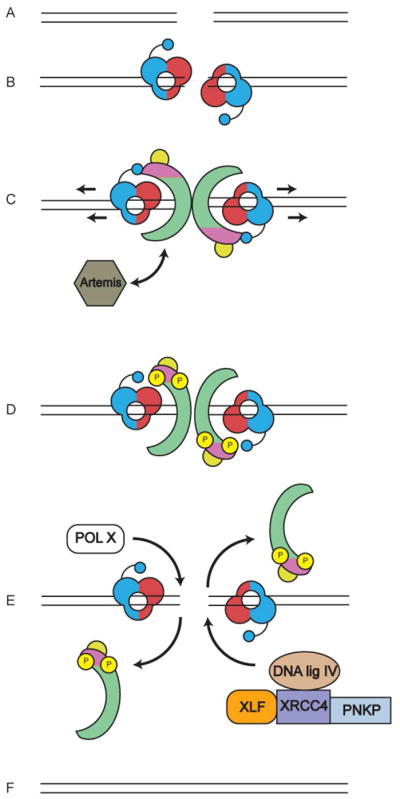
A. DSBs can be formed either by DNA damaging agents such as ionizing radiation (IR) or during the process of V(D)J recombination in the vertebrate immune system.
B. DSBs are detected by the Ku70/80 heterodimer, which binds to the ends of the DSB. Ku70 (shown in red) is positioned proximal to the DSB end and Ku80 (shown in blue) is distal to the end (Walker et al., 2001). The C-terminal domain of Ku80 is composed of a flexible linker with a globular domain and a C-terminal region that interacts directly with DNA-PKcs (Gell and Jackson, 1999), (shown as small blue circle). Significantly, SAXS revealed that this flexible domain extends towards Ku70, positioning the DNA-PKcs interacting region close to the end of the DSB (Hammel et al., 2010b). Ku plays a key role in NHEJ, being responsible for recruitment of multiple downstream proteins, including DNA-PKcs and XLF, to the DSB (reviewed in (Radhakrishnan et al., 2014; Wang and Lees-Miller, 2013)).
C. Upon activation, DNA-PKcs is recruited to the DSB end, causing Ku to move inwards (Frit et al., 2000) (indicated by arrows) such that DNA-PKcs is now positioned at the extreme end of the DSB (Yoo and Dynan, 1999). dsDNA interacts with the central region of DNA-PKcs (Boskovic et al., 2003; Williams et al., 2008). It has been suggested that DNA-PKcs may open or fray the ends of the dsDNA which may allow single stranded DNA to bind in a channel in the head domain of DNA-PKcs (DeFazio et al., 2002) (see also Figure 2B). DNA-PKcs also interacts with Artemis (shown in gray), an endonuclease that, in the presence of DNA-PKcs, facilitates DNA hairpin opening at coding joints in V(D)J recombination (Goodarzi et al., 2006). The role of Artemis in repair of IR-induced DSBs is not known. DNA-PKcs is coloured as in Figure 2A, where the N-terminal HEAT repeats are in green, the FAT and FATC domains in magenta and the kinase domain in yellow, as described by (Sibanda et al., 2010).
D. DNA-PKcs undergoes extensive autophosphorylation (yellow circles with P), which in vitro has been shown to induce large conformational changes and promote release of phosphorylated DNA-PKcs from the Ku-DNA complex (Chan and Lees-Miller, 1996; Dobbs et al., 2010; Hammel et al., 2010b). A recent study suggests that recruitment of the XRCC4-DNA ligase IV complex to the DSB is required for phosphorylation of DNA-PKcs (Cottarel et al., 2013). Artemis has also been reported to interact with DNA ligase IV (Malu et al., 2012; Ochi et al., 2013) (not shown in figure).
E. DNA-end processing enzymes such as polynucleotide kinase/phosphatase (PNKP), which interacts with XRCC4 (Koch et al., 2004) and DNA polymerases mu and/or lambda of the pol X family are also recruited to the DSB, as is the XRCC4-DNA ligase IV-XLF complex, which seals the DSB ends. XRCC4 and XLF form long helical filaments (not shown in figure) that may facilitate bridging of ends prior to ligation (Hammel et al., 2011; Hammel et al., 2010a; Mahaney et al., 2013).
F. NHEJ is completed when the processed ends of the DSB are covalently joined. How and when Ku is removed from DSB ends is not known precisely but may involve ubiquitination (Feng and Chen, 2012; Postow, 2011; Postow et al., 2008).
