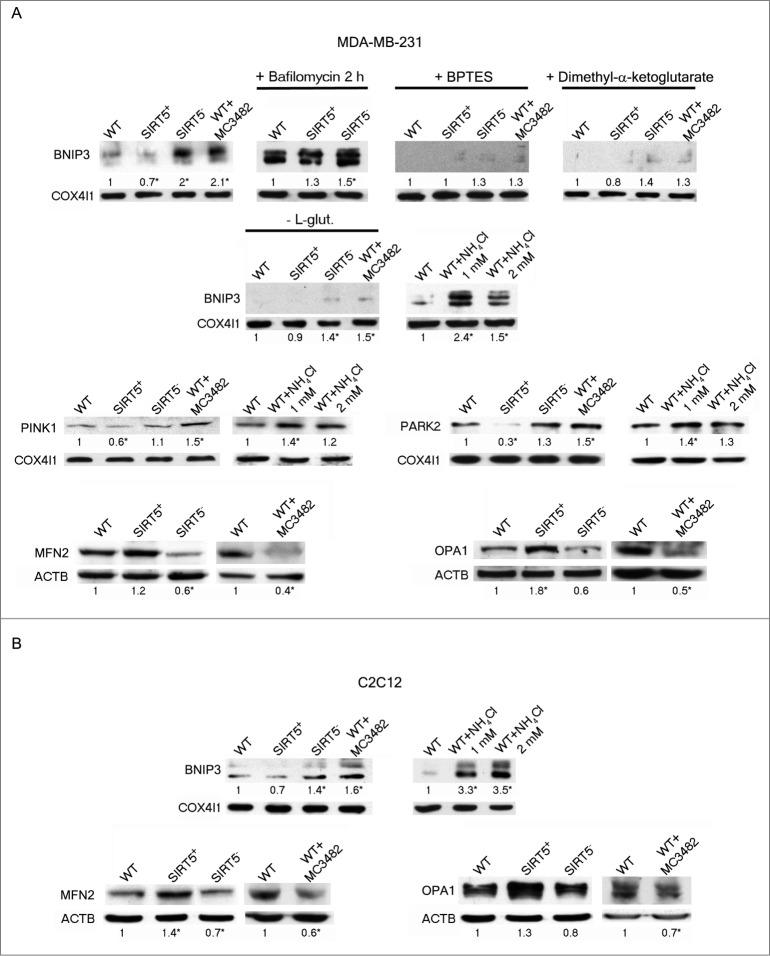
Figure 7.
SIRT5 controls ammonia-induced mitophagy and mitochondrial fusion proteins. (A) Upper panel: MDA-MB-231 WT cells in the presence or absence of MC3482, as well as SIRT5+ and SIRT5− clones were processed to obtain mitochondrial fractions. Alternatively, WT, SIRT5+ and SIRT5− cells were treated with 100 nM bafilomycinA1, BPTES, dimethyl-α-ketoglutarate, NH4Cl or grown without L-glutamine for 17 h and processed. BNIP3 expression levels were determined by western blot as indicated under Materials and Methods. Densitometric analysis of the gels was performed as described under Materials and Methods. COX4I1 was used as a loading control. *Significantly different from WT cells. Significance was set at P < 0.05. Middle panel: MDA-MB-231 WT cells in the presence or absence of MC3482, as well as SIRT5+ and SIRT5− clones were processed to obtain mitochondrial fractions. Alternatively, WT, SIRT5+ and SIRT5− cells were treated with NH4Cl for 17 h. PINK1 and PARK2 levels were assessed by western blot as indicated under Materials and Methods. Densitometric analysis of the gels was performed as described under Materials and Methods. COX4I1 was used as a loading control. *Significantly different from WT cells. Significance was set at P < 0.05. Lower panel: MDA-MB-231 WT cells in the presence or absence of MC3482, as well as SIRT5+ and SIRT5− clones were processed to obtain whole cellular extracts. MFN2 and OPA1 levels were assessed by western blot as indicated under Materials and Methods. Densitometric analysis of the gels was performed as described under Materials and Methods. ACTB was used as loading control. Data are representative of at least 3 separate experiments. *Significantly different from WT cells. Significance was set at P < 0.05. (B) Upper panel: C2C12 WT cells in the presence or absence of MC3482, as well as SIRT5+ and SIRT5− clones were processed to obtain mitochondrial fractions. Alternatively, WT, SIRT5+ and SIRT5− cells were treated with NH4Cl for 17 h. BNIP3 expression levels were determined by western blot as indicated under Materials and Methods. Densitometric analysis of the gels was performed as described under Materials and Methods. COX4I1 was used as a loading control. *Significantly different from WT cells. Significance was set at P < 0.05. Lower panel: C2C12 WT cells in the presence or absence of MC3482, as well as SIRT5+ and SIRT5− clones were processed to obtain whole cellular extracts. MFN2 and OPA1 levels were assessed by western blot as indicated under Materials and Methods. Densitometric analysis of the gels was performed as described under Materials and Methods. ACTB was used as loading control. Data are representative of at least 3 separate experiments. *Significantly different from WT cells. Significance was set at P < 0.05.