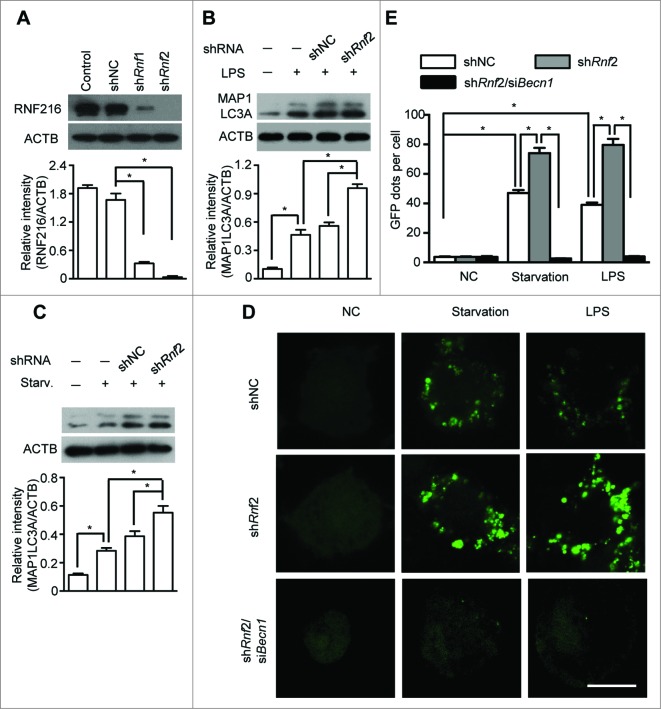
Figure 2.
Knockdown of RNF216 expression abrogates the inhibition of RNF216 on autophagy induction. (A) RAW 264.7 cells were infected with lentivirus with scrambled shRNA (shNC) or Rnf216 shRNA1 and 2 (shRnf1, and 2) (MOI=10), and 48 h later, the cells were lysed and subject to SDS-PAGE followed by being transferred to nitrocellulose membrane. After blotting with RNF216 antibody, the membrane was incubated with HRP-conjugated secondary antibody, and visualized with an ECL chemiluminescence kit. (B and C) RAW 264.7 cells infected with lentivirus with shNC or shRnf2 (MOI=10) were treated with LPS (100 ng/mL) for 16 h (B), or starvation for 4 h (C), then the cells were lysed and subjected to SDS-PAGE followed by being transferred to nitrocellulose membrane. After blotting with MAP1LC3A antibody, the membrane was incubated with HRP-conjugated secondary antibody, and visualized with an ECL chemiluminescence kit. The band densitometry was quantified using ImageJ software. The quantitative data were calculated from 3 independent experiments, and were shown as mean ± SEM. (D) RAW 264.7 cells infected with lentivirus containing scrambled shNC, or shRnf2 (MOI=10) alone or combined with siBecn1 transfection were grown on coverslips, and transiently transfected with GFP-MAP1LC3A overnight, followed by treatment with LPS (100 ng/ml) for 16 h or starvation for 4 h, and then fixed. Digital images were captured with confocal microscopy. Scale bar = 10 μm. (E) Cells with featured puncta were considered as autophagy-positive, and at least 100 cells were quantified. Puncta dots per cell were shown as mean ± SEM. (*P < 0.05).