Abstract
MicroRNA-155 plays an important role in the inflammatory response macrophages, while present studies identified that miR-155 was up-expressed in gastritis induced by Helicobacter pylori. We found that miR-155 was over expressed in macrophages infected with H. pylori in vivo or in vitro. Subsequently, inflammatory cytokines IL-23, IL-10, TNF-α and IL-8 were increased significantly, and the expression of CD80, CD86 or COX2, NOS2. Were enhanced in H. pylori infection macrophages by regulated with miR-155mimics. Furthermore, the apoptosis of macrophages induced by H. pylori was increased obviously due to the over-expression of miR-155. Therefore, these observations indicated that miR-155 may act as a inflammatory promoter in H. pylori infected macrophages. These findings contribute us to understand the functions of miR-155 in gastritis induced by H. pylori furtherly.
Keywords: Macrophage, miR-155, Helicobacter pylori
Introduction
Helicobacter pylori (H. pylori) is a gram-negative, microaerophilic bacterium that could inhabit in the stomach and cause gastritis, peptic ulcer, and gastric cancer [1]. In acute or chronic H. Pylori infection, inflammation is thought to be a major determinant of both peptic ulceration and gastric malignancy [2], but the regulatory mechanisms of H. pylori-induced inflammation are still not well understood.
Gastric epithelial cells constitute the first line of defense against H. pylori, and they produce interleukin-8 (IL-8), which promotes the recruitment of poly nuclear cells [3]. Immune cells in gastric tissures including macrophages, dendritic cells (DC) and mucosa infiltrating lymphocytes take part in the innate and adaptative immune responses to H. pylori. Host defense against pathogens requires the induction of appropriate innate immune responses, as excessive or inappropriate activation of the immune system can be deleterious to the organism. Therefore, various immune regulators, including microRNAs (miRNA), also take part in the immune responses [4].
MicroRNAs (miRNAs) are non-protein coding 20~22 nucleotide small RNAs that induce translational repression or degradation of their mRNA targets [5,6]. MiRNA received considerable attention because of their implication in maintaining homeostasis in fundamental biological processes in non-pathological states, and their deregulation in pathological states [7]. A growing body of evidence suggests bacterial and viral infection of mammalian and plant cells can modulate miRNA expression [8]. Changes in miRNA expression in response to bacterial infection have been reported, including H. pylori infection in the gastric mucosa, in gastric epithelial cells and in immune cells [9-12].
Recent reports show that during Helicobacter pylori infection, miR-155 play an negative regulatory role in the inflammatory response and highlighted the regulatory role of miR-155 in H. pylori infection and associated diseases [13]. Meanwhile, miR-155, have recently emerged as crucial regulators of innate immunity and inflammatory responses, and is regulated by Toll-like receptor (TLR) ligands in monocyte-derived cells and has been shown to be induced in macrophages during H. pylori infection [14]. Macrophages cells are key players in vivo in terms of inflammation and T or B cell lymphoma initiation. To date, however, there few studies of the regulation of miRNAs by H. pylori in macrophage cells.
Here, we investigated the function of miR-155 in the inflammatory response of H. pylori infection macrophages, and its role in the activation or apotosis of macrophages infected with H. pylori.
Materials and methods
Peripheral blood mononuclear cells collection
Blood samples were collected from 30 H. pylori infection patients and 30 healthy individuals from the Cancer Institute of ChongQing in 2014. Peripheral whole blood (5 mL) was collected in EDTA-anticoagulated tubes. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Ficoll-Paque™ Plus (Amersham Pharmacia Biotech, Shanghai, China) according to the manufacturer’s instructions. After Ficoll density gradient centrifugation, the upper layer was removed carefully, and the PBMCs were collected and washed twice with PBS by low-speed centrifugation (200× g, 10 min). PBMCs were frozen at -80°C within 4 h of blood collection. All procedures were approved by the ethics committee of the Cancer Institute of Chongqing.
Macrophage cultures
The human acute monocytic leukemia cell line THP-1 was purchased from the Center for Type Culture Collection of Wuhan University. Cells were cultured in wells or flasks at 37°C under 5% CO2, in RPMI 1640-GlutaMAX™ (HyClone Laboratories, GE Healthcare Lifesciences, Logan, UT, USA) containing 10% (v/v) fetal bovine serum (HyClone), 100 U/ml penicillin, 0.1 mg/ml streptomycin and 0.25 μg/ml amphotericin B. Differentiation of THP1 cells into macrophage-like cells was induced by stimulation with 0.1 mmol/l phorbol 12-myristate 13-acetate, PMA (Sigma, St Louis, MO, USA) for 24 h.
H. pylori culture
The wild-type H. pylori strain 26695 obtained from ATCC were cultivated for 48 h at 37°C under microaerobic conditions (5% O2) on selective agar consisting of 21.5 g of Wilkins Chalgren agar, 50 ml of human blood, 10 μg/ml of vancomycin, 10 μg/ml of cefsulodin, 5 μg/ml of trimethoprim, and 10 μg/ml of amphotericin B.
siRNA transfection
THP-1 were cultured in a 12-well plate for 24 h, the cells were washed with PBS and 500 μL OpitMEM (Naomi Biotech, WuXi, China) was added to each well of the 12-well plate. Then, 20 pmol miR-155 mimics, miR-155 inhibitors, negative siRNA, or a blank control were added to 50 μL OpitMEM and combined with 50 μL OpitMEM containing 5 μL IR that was heat shocked for 10 s at 37°C. The mixed solutions were then added to the cells. After 6 h, the medium was replaced and the cells were cultured for 24 h. The transfected cells were observed by fluorescence microscopy. Differentiation of these cells into macrophage-like cells was induced with 0.1 mmol/l Phorbol 12-myristate 13-acetate, PMA (Sigma, St Louis, MO, USA) for 24 h.
Co-cultures of macrophages and H. pylori
Immature macrophages were washed once in PBS and plated onto 24-well plastic plates at a density of 5×105 cells per well in 1 ml of RPMI-1640 growth medium. H. pylori were recovered from the agar plates using a swab and resuspended in RPMI-1640 growth medium at an optical density of 0.6 at 600 nm, which corresponds to 3×107 CFU/ml. The bacteria were added to macrophages at the indicated multiplicity of infection (MOI) 10:1 and the co-cultures were further incubated at 37°C in a 5% CO2 atmosphere for 24 h.
Quantitative RT-PCR of miR-155
Levels of miR-155 expression were analyzed by quantitative RT-PCR performed with the Eppendorf PCR system (Eppendorf, Germany). Total RNA was isolated from PBMCs or macrophages treated with LPS or H. pylori using TRIzol reagent and quantified by a photometer. cDNA was synthesized by M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
For RT-PCR, 1 μL cDNA, 0.5 μL forward primer and 0.5 μL reverse primer, 2 μL 2.5 mM dNTPs, 0.5 μL DNA polymerase (10 U/μL) (Takara, Da lian, China), 4 μL 5× buffer, and 11.5 μL dd H2O were reacted for 35 cycles at 95°C for 10 min, 95°C for 15 s, and 60°C for 1 min. A U6 small nuclear RNA was used as endogenous control for data normalization, and relative expression was calculated using the comparative threshold cycle (Ct) method. The primers were: 5’CGCTGCTACCTGAGAGTAGACCAGATGCAGCG ACCCCT3’ (reverse trans-scription), R 5’TTAATGCTAATCGTGATAG3’, and F 5’ACCTGAGAGTAGACCAGA 3’ for miR-155; R 5’GGAACGCTTCACGAATTTG3’ and F 5’ATTGGAACGATACAGAGAAGATT3’ for U6.
The expression levels of miR-155 were normalized to those of U6 miRNA. Relative expression was calculated with the 2-ΔΔCt formula: ΔΔCt = ΔCtreference - ΔCtsample, where ΔCt is the difference in the cycling threshold between the gene of interest and the housekeeping gene U6, ΔCtsample is the Ct value for miR-155 normalized to U6, and ΔCtreference is the Ct value corresponding to control samples normalized to U6. Mean Ct values were calculated from the duplicates.
Cytokine quantifications by ELISA
Macrophages cultured in six well plates were infected with H. pylori (MOI of 10; circles) for 24 h. IL-23, IL-10, TNF-α, IL-8 in above cell culture supernatants were analyzed by ELISA kits, according to the instructions of Boster Biotechnology Company (WuHan, China), and cytokine concentrations were calculated by referring to standard curves.
Flow cytometry analysis
Macrophages transfected with siRNA cultured in six well plates were treated for 24 h with H. pylori (MOI of 10; circles), then were harvested and washed twice with PBS containing 0.2% BSA. Cells were then stained with fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-labeled monoclonal antibodies (BD, USA) to CD80, CD86, or the appropriate isotype controls. Macrophages were washed and fixed in 10% (vol/vol) formaldehyde-PBS. Finally, cell sorting and analysis were performed on a flow cytometry (Beckman MOFLO XDP, USA). Median fluorescence intensities (MFIs) and the percentages of positively expressing cells were determined after subtraction of the values for the isotype controls.
Western blotting analysis
For Western blotting, equal concentration of proteins from cell lysates, as quantitated by the Micro BCA Protein Assay (Merck Millipore, USA) were loaded on 10% SDS-PAGE gels, electrophoresed, and transferred onto polyvinylidene difluoride membrane (Milipore, Bedford, MA). The membranes were probed for COX2 (R&D, 1:600 dilution), NOS2 (R&D, 1/400 dilution). Immune-detected protein bands were quantified with ImageJ and statistically analyzed by ANOVA software.
Apoptosis and necrosis
Macrophages interferenced with siRNA were infected with H. pylori (MOI of 10; circles) for 24 h. Cell apoptosis was quantitatively determined by flow cytometry using an annexin V-FITC/PI apoptosis detection kit (BD, USA). Following the treatment, cells were harvested by trypsination, washed with PBS, and incubated with annexin V-FITC and PI at 25°C for 10 min in the dark. The stained cells were analyzed by an FACS Calibur flow cytometer and CellQuest analysis software (Beckman MOFLO XDP, USA).
Statistical analysis
All data were expressed as mean ± SEM. Values were analyzed by SPSS16.0 software for Windows, and the statistical significance of difference among the values was evaluated by one-way analysis of variance. P value <0.05 was defined as significant.
Results
The expression of miR-155 in H. pylori infection patients or macrophages
To investigate the role of miR-155 in H. pylori infection cells, the expression levels of miR-155 were first measured in 30 cases of PBMCs from H. pylori infection individual by quantitative PCR (Figure 1A), then the levels of miR-155 expression in macrophages stimulated with LPS or H. pylori, miR-155 levels were detected by quantitative PCR (Figure 1B). Above results show that miR-155 expression levels were significantly higher in H. pylori infection patients than healthy individual, and were also over-expressed in macrophages stimulated with LPS or H. pylori.
Figure 1.
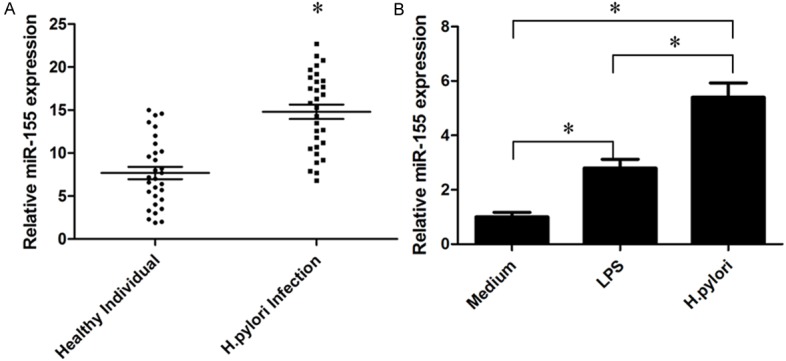
Analysis of the expression of miR-155 in PBMCs from H. pylori infection patients or H. pylori infection macrophage. A: The expression levels of miR-155 in 30 cases of PBMCs of H. pylori infection patients were detected by quantitative PCR. B: Quantitative PCR results of miR-155 expressed in H. pylori infection macrophage; U6 was used as the control (mean ± SEM, three independent experiments). *P<0.05.
Macrophages transfected with siRNAs
Macrophages cells were transfected with siRNAs including miR-155 mimics, miR-155 inhibitors, or negative control siRNA for 24 h by ImagenFect RNAi kit (Naomi Biotech, Wuxi, China). Then transfection macrophages cells were observed under a fluorescence microscope (Olympus, Osaka, Japan) to assess the transfection efficiency. Most macrophages cells emitted green fluorescence, indicating that the siRNAs were transfected into macrophages cells successfully (Figure 2).
Figure 2.
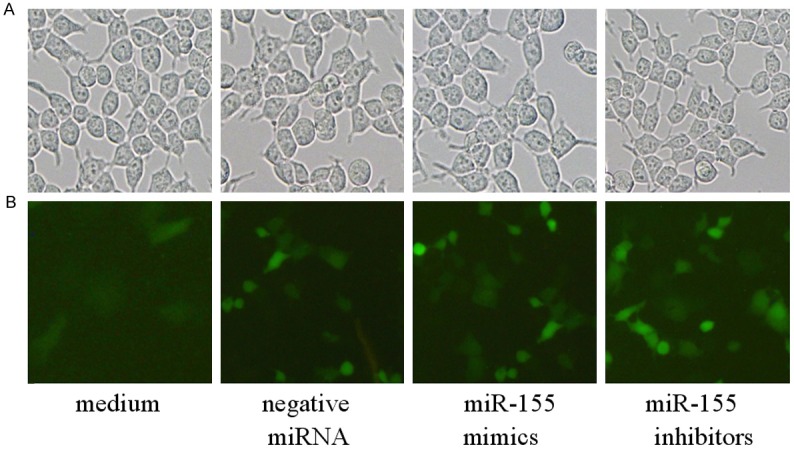
Immunofluorescence microscopy of macrophages cells transfected with siRNAs (×40). A: Untransfected macrophages cells; B: Macrophages cells transfected with siRNAs. Experiments were repeated 3 times.
Analysis of proinflammatory cytokines secreted from macrophaes
Macrophages are able to produce a variety of cytokines such as IL-6, IL-8, IFN-γ, TNF-α and so on to resist microbial invasion. Macrophages transfected with miR-155 mimics, miR-155 inhibitors, or negative control siRNA were infected with H. pylori, then IL-23, IL-10, TNF-α and IL-8 in the supernatant were detected by ELISA method (Figure 3). Notably, IL-23, IL-10, TNF-α and IL-8 were all increased significantly in H. pylori infection macrophages interferenced by miR-155 mimics, but IL-23, IL-8 were decreased in H. pylori infection macrophages treated with miR-155 inhibitors. Therefore, above results indicated that an inflammatory response of H. pylori infection macrophages could be regulated by miR-155.
Figure 3.
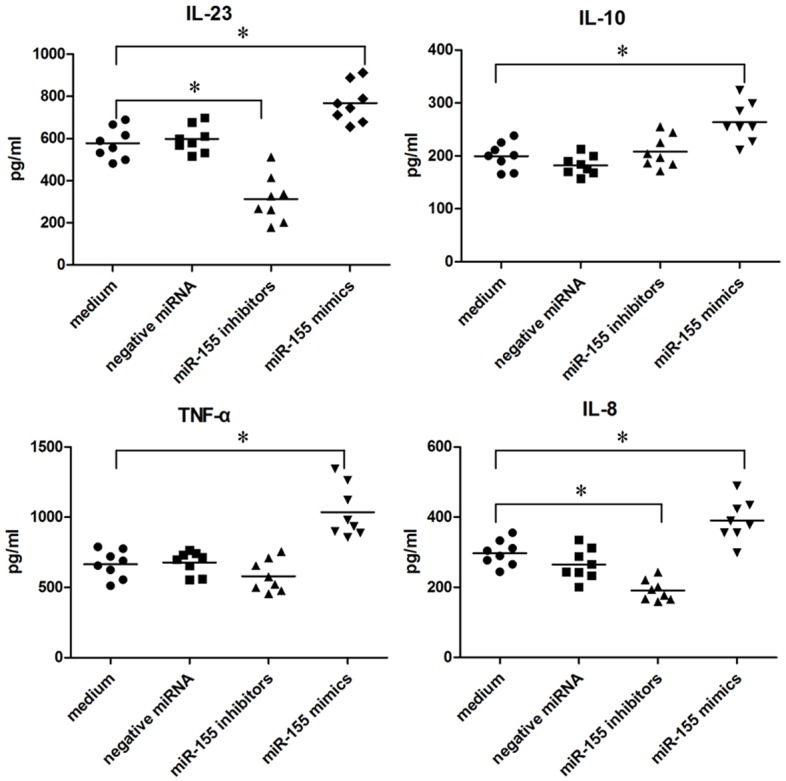
Cytokines released from H. pylori infection macrophages transfected with siRNA. A total of 106 macrophages interferenced with miR-155mimics, miR-155inhibitors or negative miRNA, were infected with H. pylori (MOI of 10; circles) for 24 h. Concentrations of cytokines in the supernatants were determined by ELISA, and each symbol per condition represents the data obtained with cells from one donor. Horizontal lines show the median values of 8 experiments. *P<0.05 (Friedman test and Dunn’s multiple comparison test).
Expression of CD80 and CD86 in siRNA interferenced macrophages infected with H. pylori (flow cytometry)
In order to confirm the enhancement of CD80/86 proteins expression by H. pylori infection in macrophages interferenced by miR-155 mimics, miR-155 inhibitors and negative miRNA, we quantified CD80/86 levels by flow cytometry. Macrophages were interferenced by miR-155 mimics, miR-155 inhibitors and negative miRNA for 24 h, and infected with H. pylori, then subjected to flow cytometry for the detection of CD80 or CD86. The results show that CD80 levels increase in miR-155 mimics transfected macrophages, but decreased in miR-155 inhibitors transfected macrophages. Meanwhile, the expression of CD86 was similar with macrophages the expression of CD80 (Figure 4).
Figure 4.
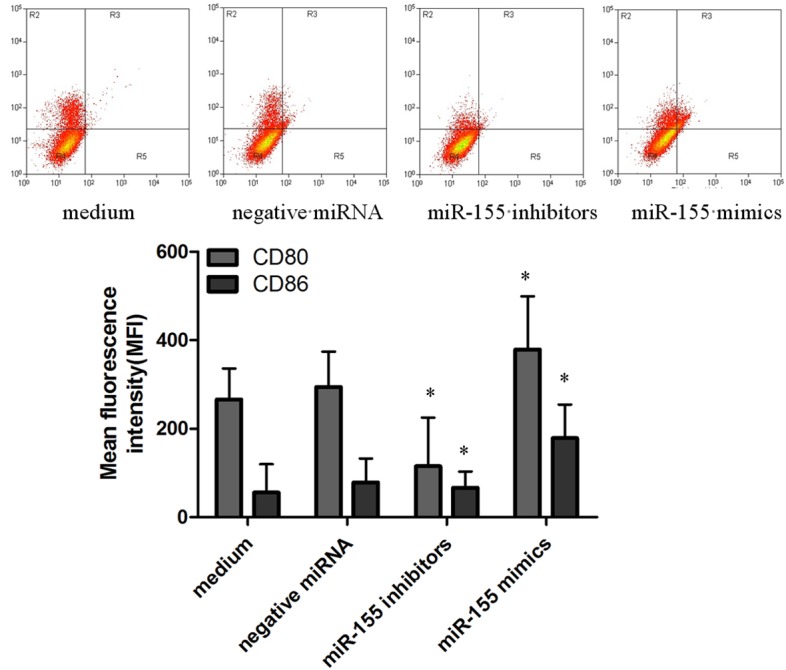
Analysis of surface CD80/CD86 by flow cytometry in siRNA interferenced macrophages infected with H. pylori. Macrophages interferenced with miR-155mimic, miR-155 inhibitors, then 106 cells were analyzed on FACS and obtained fluorescence intensities of CD80/CD86. Bar graphs show the mean fluorescence intensities (MFI) of CD80/CD86 (mean ± SE, 3 independent experiments), and statistical analysis was by SPSS 16.0, with asterisks indicating the pairs of values compared for which significant differences were observed (*P≤0.05).
Proteins expressed in macrophages
COX2, NOS2 in siRNA interferenced macrophages infected with H. pylori were analyzed by Western blotting (Figure 5), and the gray of bands was scanned by Image J software, GAPDH as standard calibration. As a result, COX2, NOS2 proteins were all decreased in miR-155 inhibitors interference macrophages, but up-expressed in miR-155 mimics interference macrophages.
Figure 5.
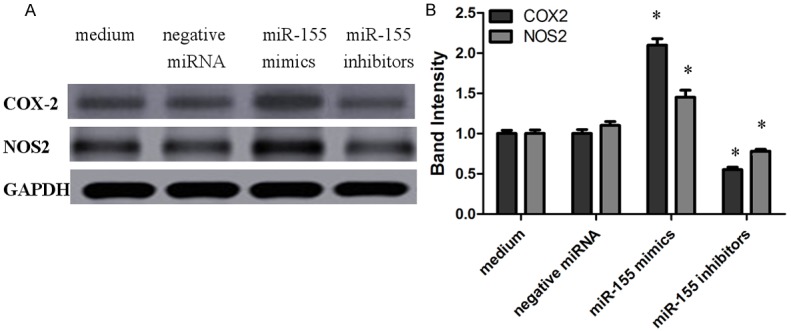
Analysis of the expression of COX2 and NOS2 proteins detected by Western blotting. COX2 and NOS2 proteins in macrophages interferenced with miR-155 mimics, miR-155 inhibitors and negative miRNA were infected with H. pylori (MOI of 10; circles), then were analyzed by Western blotting, and controls included macrophages incubated with medium alone (mean ± SEM, 3 independent experiments). Statistical analysis was by SPSS 16.0, with brackets indicating the pairs of values compared for which significant differences were observed (*P<0.045).
Apoptosis of macrophages
Macrophages interferenced by miR-155 mimics, miR-155 inhibitors and negative miRNA were incubated with H. pylori for 24 h, then were analyzed the asymmetry and permeability of the cell membrane with annexin- VFITC and PI staining by flow cytometry (Figure 6). As shown in results, the apoptosis or necrosis of macrophages interferenced by miR-155 mimics were decreased significantly compared with macrophages treated with miR-155 inhibitors, negative miRNA or medium.
Figure 6.
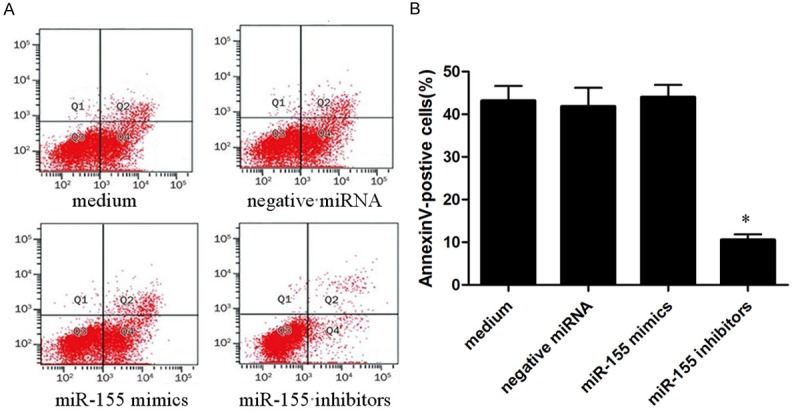
The apoptosis of siRNA interference macrophages infected with H. pylori was detected by flow cytometry. A: Results of flow cytometry, which macrophages interferenced by miR-155 mimics, miR-155 inhibitors and negative miRNA were infected with H. pylori (MOI of 10; circles) respectively for 24 h; B: Quantified assay of the apoptosis and necrosis by SPSS 16.0, with asterisks indicating the pairs of values compared for which significant differences were observed was presented as mean ± SEM (n = 3 represented for three time experiments, *P<0.05).
Discussion
Recently, miR-155 has been indicated to play a key role in the regulation of normal immunity or inflammation response [15,16]. Expression of miR-155 in monocytes or macrophages is strongly induced by a broad range of stimuli including bacterial lipopolysaccharide (LPS), IFN-β polyriboinosinic-polyribocytidylic acid (poly IC) or tumor necrosis factor-a (TNF-α) [17-19]. In addition, the up-regulated miR-155 expression has been found in synovial fibroblasts and tissue from rheumatoid arthritis patients [20]. Therefore, the mechanism by which miR-155 modulate the immune response to H. pylori is needed to be further investigated. The expression miR-155 in PBMCs of H. pylori infection patients or in macrophages infected with H. pylori was enhanced remarkerly in vivo and in vitro. Meanwhile, macrophages were transfected with miR-155 mimics, miR-155 inhibitors, or negative siRNA successfully to study functions of miR-155 in H. pylori infection macrophages.
Macrophages as myeloid antigen-presenting cells (APCs), are present in the H. pylori-infected mucosa and are likely involved in both the induction and maintenance of H. pylori specific immune responses and inflammatory reactions and have a high capacity to kill H. Pylori [21]. Macrophages could be activated by H. pylori, which the production of IL-6, IL-10 and IL-23 and the expression of CD80, CD86 were increased in macrophages infected with H. pylori [22]. In our studies, IL-23, IL-10, TNF-α and IL-8 increased in obviously in miR-155 mimics transfection macrophages infected with H. pylori, but IL-23 and IL-8 decreased in miR-155 inhibitors interference macrophages infected with H. pylori. The expression of CD80 and CD86 was enhanced in miR-155 mimics transfection macrophages infected with H. pylori, but the results was opposite in miR-155 inhibitors interference macrophages infected with H. pylori. All above results indicate that miR-155 could promote the inflammatory response through cytokine secretion and the expression of cell surface signaling molecules to kill or inhibit H. pylori.
COX-2 involved in the early inflammation in cells, but has anti-inflammatory function in chronic inflammatory stage. Animal experiments show that, in COX-2 gene knockout mouse using indomethacin could induce the gastrointestinal ulcer or peritonitis and enhanced the susceptibility of colitis. Meanwhile COX-2 could enhance the adoptive cell protective effect of gastric tissue, and involve in the regulation of gastric epithelial cell proliferation and enhance the stress resistance of mucosal injury. Proinflammatory proteins such as COX2, IL-6, IL-8, are markers of chronic inflammation in the tumor microenvironment, and these proinflammatory mediators directly correlate with inducible nitric oxide synthase (NOS2), which is an emerging biomarker of aggressive tumors that predicts poor survival in patients with elevated tumor NOS2 expression [23-27]. The over expression of miR-155 induced the expression of COX2, NOS2 proteins in H. pylori infection macrophages, while the expression of COX2, NOS2 proteins was down expressed in miR-155 inhibition macrophages infected with H. pylori.
Gram-negative bacteria may act as pro- as well as antiapoptotic factors for macrophages. H. pylori encodes both pro- and antiapoptotic effector molecules, such as CagA and VacA [28]. Primary monocyte derived cells seemed to be resistant to H.pylori-induced apoptosis [29]. The antiapoptotic potential of miR-155 has been described for several cell types, including B lymphocytes [30], and in pancreatic tumors by targeting TP53INP1 [31] and JARID2 [32]. Moreover, miR-155 has been reported to promote resistance to specific chemotherapeutics in breast cancer cells by targeting FOXO3A [33], and recent reports suggested proapoptotic potential of miR-155 through targeting SKI2 in human melanoma cells [34] or Kpc1 in murine DCs [35]. The low expression of miR-155 would be induce apotosis of macrophages infected with H. pylori, but the over expression of miR-155 has no any effect on macrophages infected with H. pylori.
Acknowledgements
This work was supported by Science and Technology Project of Jiangsu Province in China (NO: KS1425 and NO: KS1130). We thank Xiang Ya Hospital and Xiangya Medical college for technical assistance.
Disclosure of conflict of interest
None.
References
- 1.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 2.Robinson K, Argent RH, Atherton JC. The inflammatory and immune response to Helicobacter pylori infection. Best Pract Res Clin Gastroenterol. 2007;21:237–259. doi: 10.1016/j.bpg.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Hocès de la Guardia A, Staedel C, Kaafarany I, Clément A, Roubaud Baudron C, Mégraud F, Lehours P. Inflammatory cytokine and microRNA responses of primary human dendritic cells cultured with Helicobacter pylori strains. Front Microbiol. 2013;4:236. doi: 10.3389/fmicb.2013.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of posttranscriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 6.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 7.Sonkoly E, Pivarcsi A. microRNAs in inflammation. Int Rev Immunol. 2009;28:535–561. doi: 10.3109/08830180903208303. [DOI] [PubMed] [Google Scholar]
- 8.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 10.Fassi Fehri L, Koch M, Belogolova E, Khalil H, Bolz C, Kalali B, Mollenkopf HJ, Beigier-Bompadre M, Karlas A, Schneider T, Churin Y, Gerhard M, Meyer TF. Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PLoS One. 2010;5:e9500. doi: 10.1371/journal.pone.0009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belair C, Baud J, Chabas S, Sharma CM, Vogel J, Staedel C, Darfeuille F. Helicobacter pylori interferes with an embryonic stem cell micro RNA cluster to block cell cycle progression. Silence. 2011;2:7. doi: 10.1186/1758-907X-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128:361–370. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 13.Xiao B, Liu Z, Li BS, Tang B, Li W, Guo G, Shi Y, Wang F, Wu Y, Tong WD, Guo H, Mao XH, Zou QM. Induction of microRNA-155 during Helicobacter pyloriinfection and its negative regulatory role in the inflammatory response. J Infect Dis. 2009;200:916–925. doi: 10.1086/605443. [DOI] [PubMed] [Google Scholar]
- 14.Koch M, Mollenkopf HJ, Klemm U, Meyer TF. Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis. Proc Natl Acad Sci U S A. 2012;109:E1153–1162. doi: 10.1073/pnas.1116125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruggiero T, Trabucchi M, De Santa F, Zupo S, Harfe BD, McManus MT, Rosenfeld MG, Briata P, Gherzi R. LPS induces KH-type splicing regulatory protein dependent processing of microRNA- 155 precursors in macrophages. FASEB J. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 16.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 17.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/ TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 20.Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S, Kyburz D. Altered expression of microRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 21.Borlace GN, Butler RN, Brooks DA. Monocyte and macrophage killing of Helicobacter pylori: relationship to bacterial virulence factors. Helicobacter. 2008;13:380–387. doi: 10.1111/j.1523-5378.2008.00625.x. [DOI] [PubMed] [Google Scholar]
- 22.Fehlings M, Drobbe L, Moos V, Renner Viveros P, Hagen J, Beigier-Bompadre M, Pang E, Belogolova E, Churin Y, Schneider T, Meyer TF, Aebischer T, Ignatius R. Comparative analysis of the interaction of Helicobacter pylori with human dendritic cells, macrophages, and monocytes. Infect Immun. 2012;80:2724–34. doi: 10.1128/IAI.00381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glynn SA, Boersma BJ, Dorsey TH, Yi M, Yfantis HG, Ridnour LA, Martin DN, Switzer CH, Hudson RS, Wink DA, Lee DH, Stephens RM, Ambs S. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J Clin Invest. 2010;120:3843–3854. doi: 10.1172/JCI42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm EA, Ellerhorst J, Tang CH, Ekmekcioglu S. Constitutive intracellular production of iNOS and NO in human melanoma: Possible role in regulation of growth and resistance to apoptosis. Nitric Oxide. 2008;19:133–137. doi: 10.1016/j.niox.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagares-Garcia JA, Moore RA, Collier B, Heggere M, Diaz F, Qian F. Nitric oxide synthase as a marker in colorectal carcinoma. Am Surg. 2001;67:709–713. [PubMed] [Google Scholar]
- 26.Wang L, Xie K. Nitric oxide and pancreatic cancer pathogenesis, prevention, and treatment. Curr Pharm Des. 2010;16:421–427. doi: 10.2174/138161210790232194. [DOI] [PubMed] [Google Scholar]
- 27.Okayama H, Saito M, Oue N, Weiss JM, Stauffer J, Takenoshita S, Wiltrout RH, Hussain SP, Harris CC. NOS2 enhances KRAS-induced lung carcinogenesis, inflammation and microRNA-21 expression. Int J Cancer. 2013;132:9–18. doi: 10.1002/ijc.27644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oldani A, Cormont M, Hofman V, Chiozzi V, Oregioni O, Canonici A, Sciullo A, Sommi P, Fabbri A, Ricci V, Boquet P. Helicobacter pyloricounteracts the apoptotic action of its VacA toxin by injecting the CagA protein into gastric epithelial cells. PLoS Pathog. 2009;5:e1000603. doi: 10.1371/journal.ppat.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galgani M, Busiello I, Censini S, Zappacosta S, Racioppi L, Zarrilli R. Helicobacter pylori induces apoptosis of human monocytes but not monocyte-derived dendritic cells: Role of the cag pathogenicity island. Infect Immun. 2004;72:4480–4485. doi: 10.1128/IAI.72.8.4480-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J Virol. 2010;84:11670–11678. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, Chaix A, Fazli L, Motoo Y, Wang Q, Rocchi P, Russo A, Gleave M, Dagorn JC, Iovanna JL, Carrier A, Pébusque MJ, Dusetti NJ. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolisetty MT, Dy G, Tam W, Beemon KL. Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J Virol. 2009;83:12009–12017. doi: 10.1128/JVI.01182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Levati L, Alvino E, Pagani E, Arcelli D, Caporaso P, Bondanza S, Di Leva G, Ferracin M, Volinia S, Bonmassar E, Croce CM, D’Atri S. Altered expression of selected microRNAs in melanoma: Antiproliferative and proapoptotic activity of miRNA-155. Int J Oncol. 2009;35:393–400. [PubMed] [Google Scholar]
- 35.Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI, Blazar BR, Zeng Y, Zhou X. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117:4293–4303. doi: 10.1182/blood-2010-12-322503. [DOI] [PMC free article] [PubMed] [Google Scholar]


