Abstract
It has been reported that CCAT1 is involved in the development of malignancies including colon cancer and gastric cancer. However, the role of CCAT1 in HCC still remains unknown. Real-time PCR was performed to test the relative expression of CCAT1 in HCC tissues and cell lines. We performed Chi-Square Analysis to study the correlation between clinical characteristics and CCAT1 expression. Based on the correlation, cell proliferation assay, cell invasion assay, wound healing assay and cell apoptosis assay were conducted in two HCC cell lines to examine the regulatory effect of CCAT1 on the HCC cells. The results indicated that the expression of CCAT1 was significantly increased in HCC tissues and cells compared with controls. We also found that the abnormally expressed CCAT1 could promote cell proliferation, migration and invasion. Taken together, our findings demonstrated that the aberrant expression of CCAT1 promotes hepatocellular carcinoma in vitro.
Keywords: Long non-coding RNA, CCAT1, HCC, proliferation, invasion
Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer related deaths worldwide [1,2]. Because of the high incidence of chronic hepatitis B infection and liver cirrhosis, China accounts for more than half of the world’s cases each year [2-5]. Although great advances in surgical technique and chemotherapy have been achieved over the last two decades, long-term survival of HCC is still low due to the high rate of recurrence and metastasis [6,7]. Additionally, most HCC patients present with advanced metastatic disease at the initial diagnosis [8,9]. It has been reported that the development of HCC are facilitated by multiple abnormal biological processes, such as gene mutations, epigenetic alterations, and dysregulation of both coding and non-coding genes [8,10-13]. For accurate diagnosis and adequate treatment of HCC, there is a pressing need to understand the precise molecular mechanisms underlying HCC pathogenesis [14].
Based on the size, non-coding RNAs (ncRNAs) are subdivided into two major classes: small ncRNAs (< 200 nt) and long ncRNAs (lncRNAs, > 200 nt) [15,16]. Recently, microRNAs have been identified as oncogenes or tumor suppressor genes to influence biological function of cancer cells through post-transcriptional regulation of protein expression. On the contrary, lncRNAs were once thought to be transcriptional noise [17]. However, it has become increasingly clear that lncRNAs exert significant functions at various levels, including X chromosomal inactivation, chromatin remodeling, and transcriptional repression. The long non-coding RNA colon cancer associated transcript 1 (CCAT1) with abnormal expressions in many malignancies has been reported to be involved in the pathogenesis of malignant tumors including colon cancer and gastric cancer [18,19]. However, the role of CCAT1 in the development of HCC still remains elusive.
In our study, we demonstrated that CCAT1 was aberrantly increased in the HCC tissues and cells compared with the controls. The correlation between clinical characteristics and CCAT1 expression was confirmed by Chi-Square correlation analysis. Based on the correlations between clinical characteristics and CCAT1 expression, we performed in vivo experiments and demonstrated the promoting-cancer effect of CCAT1 via cell proliferation assay, cell invasion assay and wound healing assay.
Materials and methods
Clinical samples and cell culture
86 cases of patients who underwent liver cancer radical resection during December 2010 to May 2012 were recruited from the Provincial Hospital Affiliated to Shandong University (Jinan, China). Specimens were obtained immediately after surgical resection and stored at -80°C for further analysis. All patients had negative histories of exposure to either chemotherapy or radiotherapy before surgery, and there was no other co-occurrence of diagnosed cancers. This study was approved by the Ethical Committee of Provincial Hospital Affiliated to Shandong University and every patient had written informed consent.
The human HCC cell lines: L-02, HepG2, SNU423, SMMC-7721, Hep3B were obtained from the American Type Culture Collection (ATCC, USA) and 97H, 97L were purchased from Liver cancer institute, Fudan University, Shanghai. They were maintained in an atmosphere of 5% CO2 in DMEM medium (Thermo, San Jose, CA, USA) supplemented with 10% fetal bovine serum (Thermo, San Jose, CA, USA). Cell line authentication was performed by STR profiling before initiation of this project.
RNA preparation, reverse transcription and real-time PCR
Total RNA from frozen samples and cell lines was extracted by Invitrogen (Invitrogen, California, USA) according to the manufacturer’s protocol. cDNAs from all samples were synthesized from 1 μg of total RNA by PrimeScript RT Master Mix kit (Takara, Japan). The expression of CCAT1 was analyzed by real-time PCR using Quantifast SYBR Green PCR Kit (Qiagen, Dusseldorf, Germany) at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Primer sequences were as follows: CCAT1 forward primer: 5’-CATTGGGAAAGGTG-CCGAGA-3’, reverse primer: 5’-ACGCTTAGCCAT-ACAGAGCC-3’; GAPDH, forward primer: 5’-GGG-AGCCAAAAGG GTCAT-3’, and reverse primer: 5’-GAGTCCTTCCACG ATACCAA-3’. Data analyses for the gene expression were performed using the 2-ΔΔCt method.
Gene silencing of CCAT1 in cells
To knockdown the CCAT1 expression, we purchased siRNA from RioboBio (Guangzhou, China). The negative control hairpin siRNA with no sequence homology to human genes provided by the same manufacturer was used as the negative control. A total of 1.5 × 105 cells were seeded in 35 mm tissue culture dishes for 24 h, followed by transfection with 2 μg of each respective plasmid with Lipofectamine 2000 Reagent (Invitrogen, California, USA) for 24 h. Transfection efficiency was monitored by real time-PCR. The cells were then subjected to RNA extraction or functional assays.
Cell proliferation assays
HCC cells proliferation was monitored using Cell Proliferation Reagent Kit I (MTT) (Roche Applied Science). siRNA-CCAT1 and empty vector-transfected HepG2 and SNU423 cells (3,000/well) were allowed to grow in 96-well plates. Cell proliferation was measured every 24 h following the manufacturer’s protocol.
Cell apoptosis assay
Cell apoptosis was measured according to the manufacturer’s instructions using an Annexin V-FITC Apoptosis Detection kit (KeyGen Biotech, Shanghai, China), and apoptosis rates were analyzed using a flow cytometer (FACS Calibur, BD Biosciences, San Jose, USA).
Wound healing assay
Cells were seeded at 4 × 104 cells/cm2. At day 3, a straight scratch was made with a 200 µL pipette tip and the wound was photographed under the microscope. After 48 h, cells were photographed under the microscope and the area of the remaining scratch was calculated.
Cell invasion assay
A cell suspension of in 0.2 ml RPMI-1640 medium with 5% FBS was seeded into each well of the upper transwell chamber (8 μm pore size, Corning Costar Corp, USA), which was pre-coated with Matrigel. In the lower chamber, 0.6 ml RPMI 1640 with 20% FBS was added. After incubating for 28 h 37°C in a humidified incubator with 5% CO2, chambers were disassembled and the membranes were stained with 2% crystal violet for 10 min and placed on a glass slide. The number of cells penetrating across membrane was counted under a microscope in ten random visual fields.
Statistical methods
All experiments were independently repeated at least triplicate. Data were expressed as mean ± SEM. Differences between two independent groups were tested with the student’s test. All statistical analyses were carried out using SPSS version 18.0 and presented with Graph pad prism 5.0 software. The results were considered to be statistically significant at P < 0.05.
Results
Expression of CCAT1 was up-regulated in the primary HCC tissues and human HCC cell lines
To explore the expression pattern of CCAT1 in the tissues of patients with hepatocellular carcinoma, we performed real-time PCR assay. The results indicated that the basal levels of CCAT1 in 86 pairs of human HCC tissues were obviously up-regulated compared with the paratumor tissues (Figure 1A). CCAT1 expression in the six human HCC cell lines: HepG2, SNU423, SMMC-7721, 97H, 97L, Hep3B was then studied. Similarly, CCAT1 expression of HCC cell lines was up-regulated in comparison with human L02 normal liver cells (Figure 1B). To examine the correlations between CCAT1 and clinical characteristics, all HCC samples were divided into CCAT1 high-expression group (n = 43) and low-expression group (n = 43) via using the median value as cut-off. The correlations between CCAT1 expression and the clinicopathological characteristics are presented. No positive correlation with age, gender, HbeAg, cirrhosis, ALT, AFP, however, significantly correlations with tumor size (P = 0.004) and Edmondson grade (P = 0.000) were achieved (Table 1).
Figure 1.
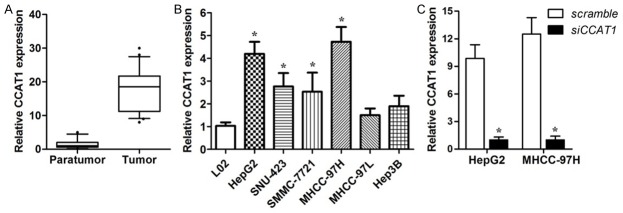
CCAT1 is up-regulated in HCC tissues and cell lines. A. The expression levels of CCAT1 in human HCC tumor tissues and paratumor tissues relative to GAPDH were determined by real-time PCR (n = 86, P < 0.05). B. CCAT1 expression levels in HCC cell lines were verified by real-time PCR. Data are represented as mean ± SEM. *Indicates P < 0.05, independent experiment was performed three times. C. CCAT1 expression levels in HepG2 and MHCC-97H cell lines transfected with CCAT1 siRNA were verified by real-time PCR. Data are represented as mean ± SEM. *Indicates P < 0.05, independent experiment was performed three times.
Table 1.
Correlation between CCAT1 expression and clinicopathological characteristics of HCC patients (n = 86)
| Characteristics | All patients | CCAT1 low expression (< Mediana) | CCAT1 high expression (≥ Mediana) | P value Chi-squared test |
|---|---|---|---|---|
| No. | 86 | 43 | 43 | |
| Age (years) | 0.194 | |||
| < 60 | 67 | 31 | 36 | |
| ≥ 60 | 19 | 12 | 7 | |
| Gender | 0.417 | |||
| Male | 69 | 36 | 33 | |
| Female | 17 | 7 | 10 | |
| HbeAg | 0.820 | |||
| Negative | 29 | 14 | 15 | |
| Positive | 57 | 29 | 28 | |
| Cirrhosis | 0.639 | |||
| Absent | 26 | 12 | 14 | |
| Present | 60 | 31 | 29 | |
| ALT (U/L) | 0.514 | |||
| ≤ 45 | 49 | 23 | 26 | |
| > 45 | 37 | 20 | 17 | |
| AFP (ng/ml) | 0.763 | |||
| ≤ 13.6 | 13 | 6 | 7 | |
| > 13.6 | 73 | 37 | 36 | |
| Tumor size (cm) | 0.004* | |||
| ≤ 5 | 31 | 22 | 9 | |
| > 5 | 55 | 21 | 34 | |
| Vascular invasion | 0.725 | |||
| Absent | 77 | 38 | 39 | |
| Present | 9 | 5 | 4 | |
| Edmondson grade | 0.000* | |||
| I+II | 62 | 39 | 23 | |
| III+IV | 24 | 4 | 20 |
The median expression level of CCAT1 was used as the cutoff.
Indicates P value < 0.05.
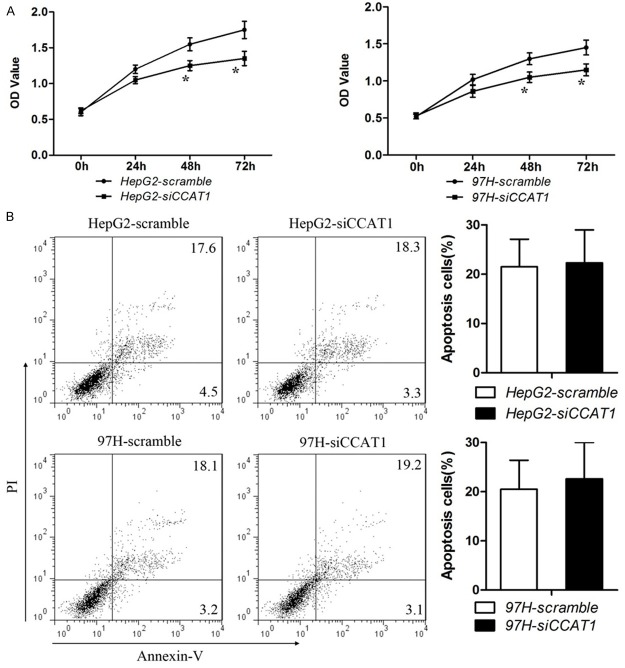
CCAT1 promoted proliferation but have no effect on apoptosis
The significant correlation between CCAT1 expression and clinicopathological characteristics suggested that CCAT1 exert a vital effect on the development of HCC. Based on the expression pattern of CCAT1, the effects of suppression of CCAT1 on cell proliferation, apoptosis were investigated in human HCC cell lines: HepG2 and 97H. The cell lines were transfected with siRNA which could inhibit the expression of CCAT1 (Figure 1C). The results showed that down-regulation of CCAT1 inhibited the activity of proliferation compared with negative control (Figure 2A). But down-regulation of CCAT1 had no effect on the apoptosis in HCC cells (Figure 2B) which is consistent with the previous studies.
Figure 2.
Effects of CCAT1 on proliferation and apoptosis in HCC cell lines. A. CCK-8 cell proliferation assays show that CCAT1 knock-down significantly weakened proliferation in HepG2 and MHCC-97H cells. Data are represented as mean ± SEM. *Indicates P < 0.05, independent experiment was performed three times. B. Representative flow cytometric plots of cell apoptosis. Cells were stained with both Annexin V and PI before analysis by flow cytometry. Numbers represent the frequency in each quadrant. Data are represented as mean ± SEM. *Indicates P < 0.05, independent experiment was performed three times.
CCAT1 promoted migration and invasion in two HCC cell experiments
We then perform in vivo experiments to explore whether CCAT1 affects migration and invasion of the two HCC cell lines. The results showed that down-regulation of CCAT1 inhibited the migration ability in cells (Figure 3A) and inhibited CCAT1 also suppressed invasion ability in the cells (Figure 3B). In other words, CCAT1 promoted migration and invasion in the two HCC cell experiments.
Figure 3.
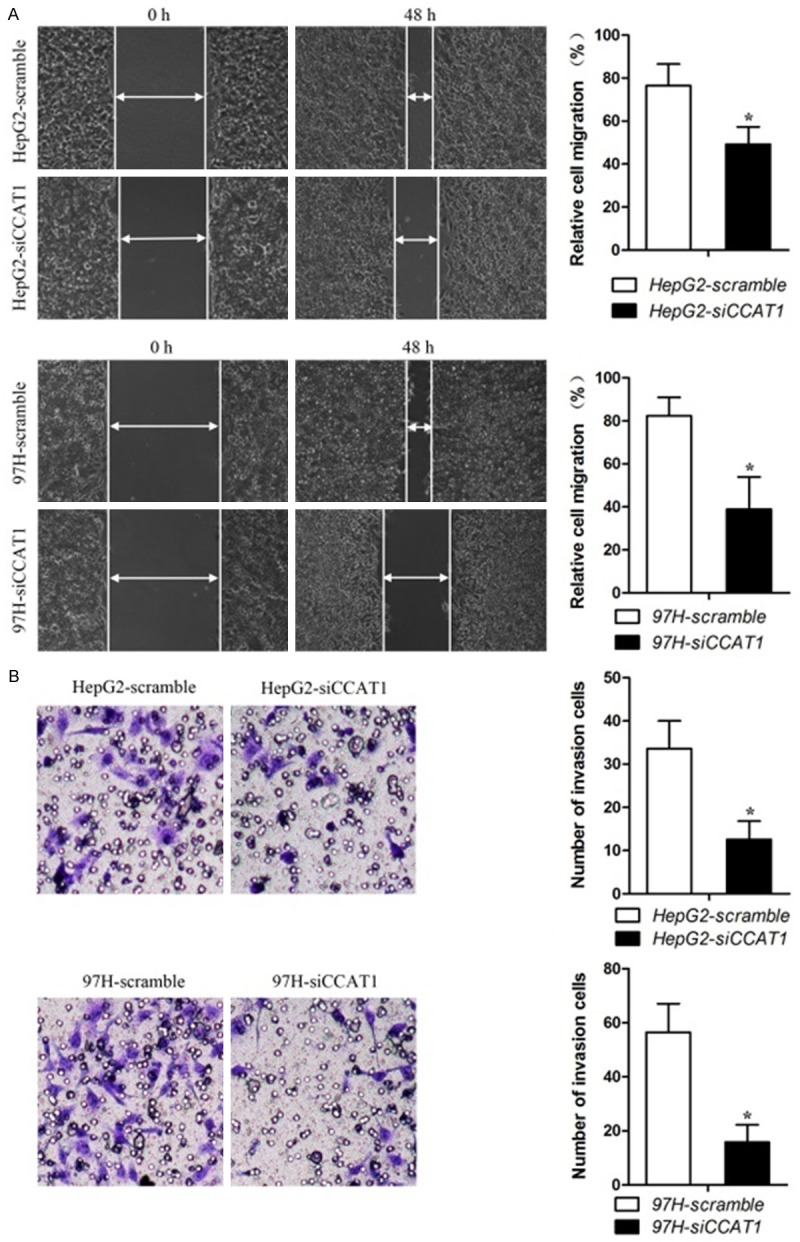
Effects of CCAT1 on migration and invasion capability in HCC cells. A. Knock-down of CCAT1 attenuated the migration ability of HCC cells. The quantifications of cell migration were presented by the histogram. All experiments were performed in triplicate and presented as mean ± SEM. *Indicates significant difference compared with control group (P < 0.05). B. Transwell chamber with matrigel assay indicated that knock-down of CCAT1 weakened HepG2 and MHCC-97H cells invasion. The quantifications of cell invasion were presented by the column chart. All experiments were performed in triplicate and presented as mean ± SEM. *Indicates significant difference compared with control group (P < 0.05).
Discussion
Long noncoding RNAs represent a novel class of noncoding RNAs that are longer than 200 nucleotides without protein-coding potential. Recently, lncRNAs were found to be dysregulated and involved in various cancer biological processes, such as proliferation, apoptosis, mobility, and invasion [17,20-25]. However, the role and precise molecular mechanism of lncRNAs in cancer development and progression still remain elusive [26-31]. In the previous study, CCAT1 was found to be involved in the development and progression of colon cancer and gastric cancer, but the functional effect of CCAT on development of HCC has not been reported.
In our study, we demonstrated that CCAT1 was aberrantly increased in tissues of patients with hepatocellular carcinoma compared with the corresponding non-tumor tissues. And the expression of CCAT1 in HepG2 and 97H cells further confirmed the above results. Additionally, Chi-Square correlation analysis was performed to demonstrate the correlation between CCAT1 expression levels and clinical characteristics. Due to the positive correlation between CCAT1 and clinical characteristics, we also performed cell proliferation assay, cell invasion assay, wound healing assay and apoptosis assay. The results suggested that down-regulation of CCAT1 suppressed cell proliferation, migration and invasion, which meant that CCAT1 could promote hepatocellular carcinoma. The results we obtained were consistent with that of He et al.[19] and Huang et al.[20], which may indicated that CCAT1 do have a vital role in the founcational regulation of many magligancies.
In summary, our findings suggested that CCAT1 was un-normally increased in the HCC tissues and cells compared with the controls. The correlation between clinical characteristics and CCAT1 expression was proved by Chi-Square correlation analysis. Based on the correlation between clinical characteristics and CCAT1 expression, we also demonstrated the promoting-cancer effect of CCAT1 via cell proliferation assay, cell invasion assay and wound healing assay. Taken together, our findings demonstrated that CCAT1 promotes HCC in vivo.
Acknowledgements
This study was supported by Natural Science Foundation of Shandong Province in China (Y2008C22 and ZR2014HM099), Excellent Youth Scientist Foundation of Shandong Province in China (2007BS03038), and others of Shandong Province (2014WS0093, 2014GGB14041, 2014WS0095 and 2014WS0096).
Disclosure of conflict of interest
None.
References
- 1.Cai JB, Shi GM, Dong ZR, Ke AW, Ma HH, Gao Q, Shen ZZ, Huang XY, Chen H, Yu DD, Liu LX, Zhang PF, Zhang C, Hu MY, Yang LX, Shi YH, Wang XY, Ding ZB, Qiu SJ, Sun HC, Zhou J, Shi YG, Fan J. USP7 accelerates p14 degradation by deubiquitinating TRIP12 and promotes HCC progression. Hepatology. 2015;61:1603–14. doi: 10.1002/hep.27682. [DOI] [PubMed] [Google Scholar]
- 2.Vigano L, Conci S, Cescon M, Fava C, Capelli P, D’Errico A, Torzilli G, Tommaso LD, Giuliante F, Vecchio FM, Salizzoni M, David E, Pinna AD, Guglielmi A, Capussotti L. Liver resection for hepatocellular carcinoma in patients with metabolic syndrome: A multicenter matched analysis with HCV-related HCC. J Hepatol. 2015 doi: 10.1016/j.jhep.2015.01.024. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Bolos D, Finn RS. Systemic therapy in HCC: lessons from brivanib. J Hepatol. 2014;61:947–950. doi: 10.1016/j.jhep.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 4.Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, Wu D, Liu L. The Long Intergenic Noncoding RNA UFC1, a Target of MicroRNA 34a, Interacts With the mRNA Stabilizing Protein HuR to Increase Levels of beta-Catenin in HCC Cells. Gastroenterology. 2015;148:415–426. e418. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Chaiteerakij R, Roberts LR. Genetic alterations in advanced HBV-related HCC with portal vein tumor thrombosis: insights from next generation DNA sequencing. J Hepatol. 2013;58:1042–1044. doi: 10.1016/j.jhep.2012.11.046. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Ramjiawan RR, Reiberger T, Ng MR, Hato T, Huang Y, Ochiai H, Kitahara S, Unan EC, Reddy TP, Fan C, Huang P, Bardeesy N, Zhu AX, Jain RK, Duda DG. CXCR4 inhibition in tumor microenvironment facilitates anti-PD-1 immunotherapy in sorafenib-treated HCC in mice. Hepatology. 2015;61:1591–602. doi: 10.1002/hep.27665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichenmuller M, Trippel F, Kreuder M, Beck A, Schwarzmayr T, Haberle B, Cairo S, Leuschner I, von Schweinitz D, Strom TM, Kappler R. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J Hepatol. 2014;61:1312–1320. doi: 10.1016/j.jhep.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Carr BI, Guerra V. HCC and its microenvironment. Hepatogastroenterology. 2013;60:1433–1437. doi: 10.5754/hge121028. [DOI] [PubMed] [Google Scholar]
- 9.Cauchy F, Fuks D, Belghiti J. HCC: current surgical treatment concepts. Langenbecks Arch Surg. 2012;397:681–695. doi: 10.1007/s00423-012-0911-2. [DOI] [PubMed] [Google Scholar]
- 10.Blum HE. Hepatocellular carcinoma: HCC. Hepat Mon. 2011;11:69–70. [PMC free article] [PubMed] [Google Scholar]
- 11.Bolondi L, Gramantieri L. From liver cirrhosis to HCC. Intern Emerg Med. 2011;6(Suppl 1):93–98. doi: 10.1007/s11739-011-0682-8. [DOI] [PubMed] [Google Scholar]
- 12.Chavez M, Danniel J, Miick R, Jarrar D, Khanmoradi K, Ortiz J. Bone Metastases From HCC. JAMA Surg. 2013;148:203–204. doi: 10.1001/jamasurgery.2013.417b. [DOI] [PubMed] [Google Scholar]
- 13.Colombo M, Iavarone M. Role of antiviral treatment for HCC prevention. Best Pract Res Clin Gastroenterol. 2014;28:771–781. doi: 10.1016/j.bpg.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 14.de Lope CR, Tremosini S, Forner A, Reig M, Bruix J. Management of HCC. J Hepatol. 2012;56(Suppl 1):S75–87. doi: 10.1016/S0168-8278(12)60009-9. [DOI] [PubMed] [Google Scholar]
- 15.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Z, Liu X, Liu L, Deng H, Zhang J, Xu Q, Cen B, Ji A. Regulation of lncRNA expression. Cell Mol Biol Lett. 2014;19:561–575. doi: 10.2478/s11658-014-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathieu EL, Belhocine M, Dao LT, Puthier D, Spicuglia S. Functions of lncRNA in development and diseases. Med Sci (Paris) 2014;30:790–796. doi: 10.1051/medsci/20143008018. [DOI] [PubMed] [Google Scholar]
- 18.Alaiyan B, Ilyayev N, Stojadinovic A, Izadjoo M, Roistacher M, Pavlov V, Tzivin V, Halle D, Pan H, Trink B, Gure AO, Nissan A. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer. 2013;13:196. doi: 10.1186/1471-2407-13-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X, Tan X, Wang X, Jin H, Liu L, Ma L, Yu H, Fan Z. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35:12181–12188. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]
- 20.Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, Zhou WP, Sun SH. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 21.Im JH, Muschel RJ. New evidence of lncRNA role in tumor progression and metastasis. Hepatobiliary Surg Nutr. 2012;1:55–56. doi: 10.3978/j.issn.2304-3881.2012.10.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, Aumayr K, Pasierbek P, Barlow DP. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 23.Maass PG, Rump A, Schulz H, Stricker S, Schulze L, Platzer K, Aydin A, Tinschert S, Goldring MB, Luft FC, Bahring S. A misplaced lncRNA causes brachydactyly in humans. J Clin Invest. 2012;122:3990–4002. doi: 10.1172/JCI65508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uva P, Da Sacco L, Del Corno M, Baldassarre A, Sestili P, Orsini M, Palma A, Gessani S, Masotti A. Rat mir-155 generated from the lncRNA Bic is ‘hidden’ in the alternate genomic assembly and reveals the existence of novel mammalian miRNAs and clusters. RNA. 2013;19:365–379. doi: 10.1261/rna.035394.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin Z, Guan D, Fan Q, Su J, Zheng W, Ma W, Ke C. lncRNA expression signatures in response to enterovirus 71 infection. Biochem Biophys Res Commun. 2013;430:629–633. doi: 10.1016/j.bbrc.2012.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng K, Guo X, Wang H, Xia J. The lncRNA-MYC regulatory network in cancer. Tumour Biol. 2014;35:9497–9503. doi: 10.1007/s13277-014-2511-y. [DOI] [PubMed] [Google Scholar]
- 27.Deshpande R, O’Reilly D, Sherlock D. Improving Outcomes with Surgical Resection and Other Ablative Therapies in HCC. Int J Hepatol. 2011;2011:686074. doi: 10.4061/2011/686074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhar D, Seki E, Karin M. NCOA5, IL-6, type 2 diabetes, and HCC: The deadly quartet. Cell Metab. 2014;19:6–7. doi: 10.1016/j.cmet.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Carlo I, Toro A. The quality of life after surgery for HCC can support the choice of this treatment. World J Surg. 2014;38:3035. doi: 10.1007/s00268-014-2677-x. [DOI] [PubMed] [Google Scholar]
- 30.Di Martino M, Di Miscio R, De Filippis G, Lombardo CV, Saba L, Geiger D, Catalano C. Detection of small (≤2 cm) HCC in cirrhotic patients: added value of diffusion MR-imaging. Abdom Imaging. 2013;38:1254–1262. doi: 10.1007/s00261-013-0009-5. [DOI] [PubMed] [Google Scholar]
- 31.Doerr A. Unraveling the lncRNA mystery. Nat Methods. 2014;11:890. doi: 10.1038/nmeth.3089. [DOI] [PubMed] [Google Scholar]