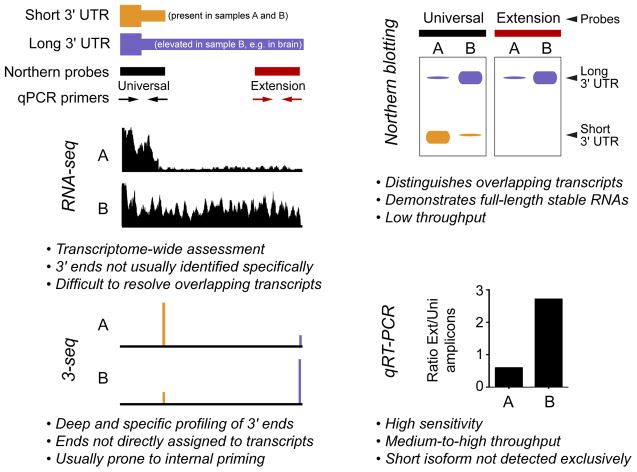
Figure 2. Experimental approaches to study alternative polyadenylation.
The schematics at top left depict a model gene that expresses short (orange) and long (purple) 3′ UTR isoforms, of which the latter is preferentially expressed in sample B and not in sample A. This pattern is representative of many neural-restricted 3′ UTR extensions of genes that are expressed more broadly. These APA isoforms can be analyzed at the level of individual genes using Northern probes or RT-PCR primers directed at proximal and distal regions of the 3′ UTR; the former is “universal” since it detects both isoforms, whereas the “extension” reagents specifically recognize the long 3′ UTR. The right panels illustrate model data for Northern and qPCR measurements of APA isoforms. The middle and bottom left panels illustrate genomewide approaches, and how model data appear at specific genomic loci. RNA-seq data provides an overview of the transcriptome as reconstructed from short overlapping reads, whereas 3′-seq methods are designed to specifically capture and sequence the polyadenylated 3′ termini of mRNAs.