Fig. 6. Sequencing the evolved lineages.
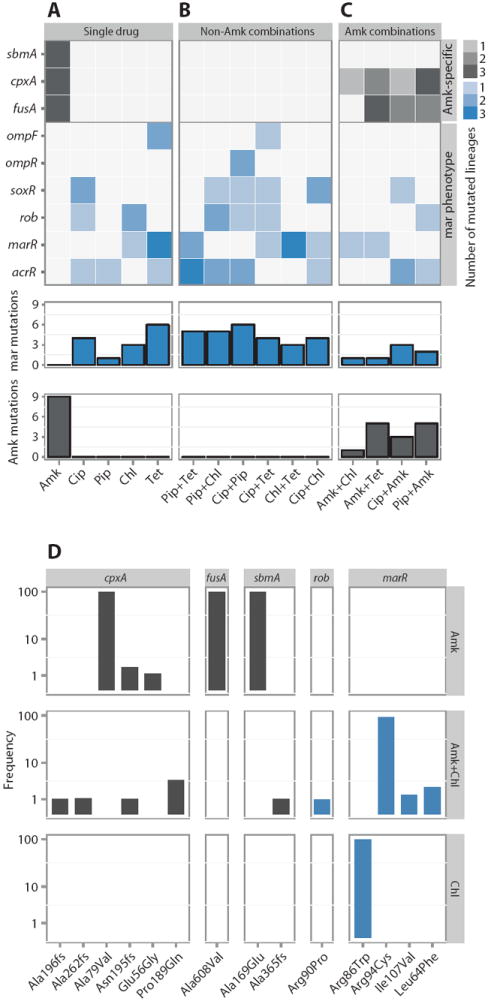
(A to C) Heat map depicting mutations (SNPs and INDELs) in the 45-endpoint sequenced lineages known to be involved in Amk and mar resistance. Mutations in the single-drug evolved lineages (A), mutations found in lineages evolved to drug pairs without Amk (B), and lineages evolved to drug pairs with Amk (C). The three parallel-evolved lineages are collapsed by drug condition. The gene targets are grouped by the phenotypic characteristics (gray, Amk-specific mutations; blue, mutations known to confer the mar phenotype). The legend indicates the number of parallel lineages that contained a mutation in the specific target. The bars below each drug condition summarizes the mar- and Amk-specific mutations, respectively. (D) Population frequency sequencing. Total DNA from the populations evolved to Amk, Chl, and Amk + Chl (drug conditions are noted in the vertical strip text) was sequenced, and frequencies of the individual SNPs/INDELs were calculated (loci are noted in the horizontal strip text). SNPs/INDELs in genes linked to Amk resistance (fusA, sbmA, and cpxA) were fixed in the Amk-evolved population but could not be fixed when Chl was present. In contrast, SNPs in marR were observed in both the Chl and Amk + Chl evolved lineages. These findings corroborate the single-isolate sequencing results.
