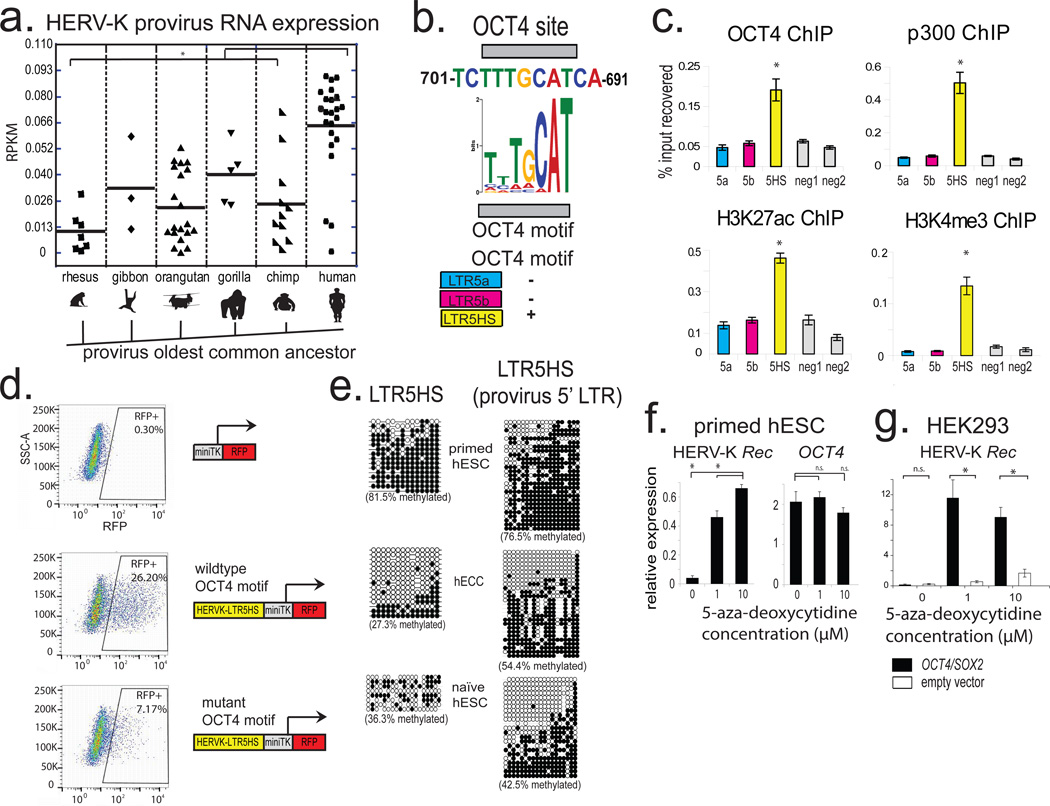
Figure 2. Transactivation by OCT4 and DNA hypomethylation of LTR5HS synergistically regulate HERV-K transcription.
a) Expression of different HERV-K proviral sequences, grouped according to the oldest common ancestor, as defined by Subramanian et al. 2011. * denotes p-value <0.05, non-paired Wilcoxon test. Solid line = mean. RNA-seq dataset used for the analysis was from 3iL naïve H1 cells (Chan et al. 2013); n= 3 biological replicates.
b) Conserved OCT4 site in LTR5HS with position weight matrix of the corresponding motif shown for comparison (top). Presence/absence of OCT4 motif in distinct LTR5 sequences is indicated (bottom); more detailed sequence information in Extended Data Fig. 2a.
c) ChIP-qPCR analyses from hECCs (NCCIT) using antibodies indicated on top of each graph. Signals were quantified using primer sets specific to LTR5HS, LTR5a, and LTR5b consensus sequences or two “negative” intergenic, non-repetitive regions. * denotes p-value <0.05 compared to negative control, one sided t-test, n=4 biological replicates, error bars are +/− 1 S.D.
d) Flow cytometry analysis of hECCs with integrated LTR5HS fluorescent reporters, either wild type (middle) or with OCT4 motif mutation (bottom). RFP positive population was gated using side-scatter area (SSC-A) and cells with integrated negative control reporter (top). Shown is a representative result of two independent experiments.
e) Bisulfite conversion quantification of LTR5HS 5-methyl-cytosine levels measured using LTR5HS-specific primer pairs anchored in the LTR5HS consensus sequence (left) or provirus-specific 5' LTR5HS (right) for hECCs (NCCIT) or hESCs (H9) or naïve hESC (ELF1). Filled circles depict modified cytosines, empty circles depict unmodified cytosines. hECC (NCCIT) and naïve hESC (ELF1) are less methylated than hESC (H9), p <0.05, non-paired Wilcoxon test.
f) RT-qPCR analysis of hESC (H9) treated with indicated concentrations of 5-aza-2'-deoxycytidine for 24 hours. *denotes p-value <0.05, one-sided t-test, n=3 biological replicates, error bars +/− 1 SD.
g) RT-qPCR analysis of HERV-K Rec RNA levels in HEK293 cells treated with indicated concentrations of 5-aza-2'-deoxycytidine, followed by transfection with OCT4/SOX2 expression constructs. *denotes p-value <0.05, one-sided t-test, n=4 biological replicates, error bars +/− 1 S.D.