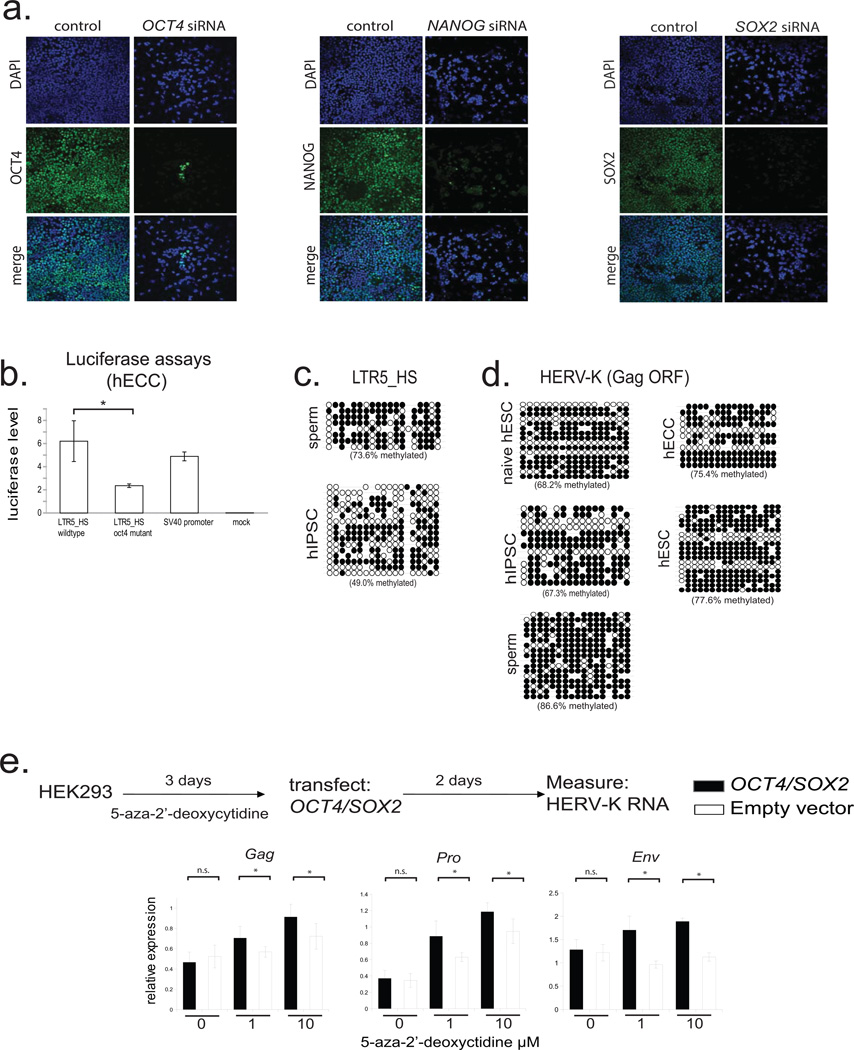
Extended Data Figure 3. HERV-K regulation by OCT4 and DNA methylation (supporting Fig. 2).
a) Transcription factor knockdown in hECCs (NCCIT). Cells were transfected with siRNA pools targeting indicated TFs and protein depletion was measured by immunofluoresence with respective antibodies in comparison to control, mock-transfected cells. DAPI (blue), OCT4 (green, left), NANOG (green, middle), SOX2 (green, right). Shown is one of three representative fields of view.
b) Dual luciferase assays with indicated reporter constructs in hECCs (NCCIT) showing that mutation of OCT4 site decreases reporter activity. N=3 biological replicates, error-bars = +/− 1 S.D.* = p-value <0.05, one-sided t-test. SV40 enhancer/promoter construct was used as a positive control.
c) Bisulfite sequencing for indicated cell types (WT33 hIPSC) analyzing consensus LTR5HS-specific amplicon as in Fig. 2f.
d) Bisulfite sequencing analysis of HERV-K proviral consensus amplicon containing 3' end of LTR, primer binding site, and 5' region of Gag ORF (see Extended Data Fig. 2a) in indicated cell types: ELF1 naïve, hESC, WT33 hIPSC, NCCIT hECC, or H9 hESC.
e) RT-qPCR analysis of HERV-K RNA levels in HEK293 cells treated with indicated concentrations of 5-aza-2'-deoxycytidine for three days, followed by transfection with OCT4/SOX2 expression constructs and RNA collection 48h after transfection. qPCR primer sets designed to 3 independent amplicons of HERV-K. *denotes p-value <0.05, one-sided t-test, n=4 biological replicates, error bars +/− 1 SD.