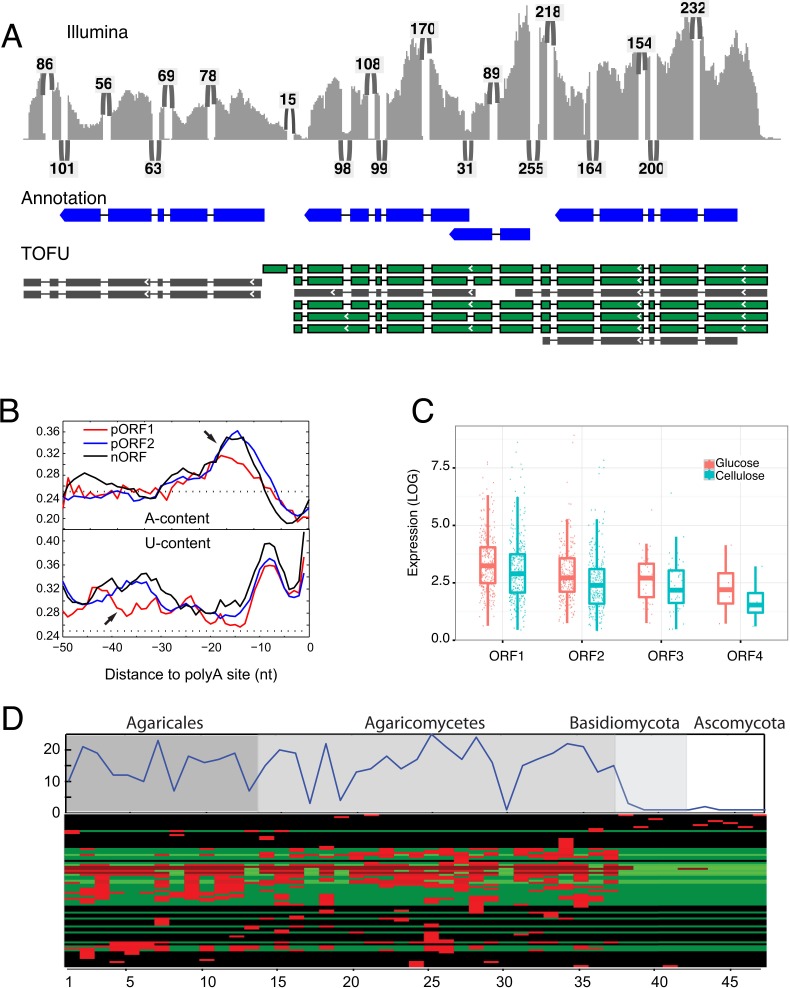
Fig 4. The genome-wide presence of polycistronic mRNAs.
a, Short-reads (Illumina) aligned to a cluster of tandem reference genes (Annotation, 3 tandem genes on the first row). The numbers of supporting short-reads for each junction are indicated. Polycistronic transcripts (TOFU) are shown in green and non-polycistronic transcripts in gray. b, A comparison of transcription termination signals. The sequence composition profiles (upper panel for A-content and lower panel for U-content) before the polyadenylation sites for different classes of ORFs. pORF1 is the upstream ORF and pORF2 is downstream ORF, while nORF stands for non-polycistronic mRNAs. The y-axis are the frequencies of a specific nucleotide averaged for 200 randomly sampled polycistronic mRNA or non-polycistronic controls, dotted lines are the expected frequencies (0.25) if all four bases are equally likely. Arrows denote NUE (upper panel) and FUE (lower panel), respectively. For this figure, only polycistronic transcripts with exactly two ORFs are plotted. Genome-wide analysis base composition of termination signals for all transcribed loci is shown in Fig B in S2 File c, The independent expression levels of ORFs within polycistronic RNAs. ORF numbers indicate their order in the transcript (5’- to 3’). d, Polycistronic transcripts are likely a unique feature to Agaricomycetes. The top plot shows the total number of adjacent ORF pairs within polycistronic transcripts from P. crispa that have conserved gene configuration in related species. The numbers on x-axis are species with increasing evolutionary distance. The bottom heatmap shows the conservation for each individual pair of ORFs. Red indicates the presence of a homologous gene pair in the species.