Abstract
Background
Rice endosperm is composed of aleurone cells in the outermost layers and starchy endosperm cells in the inner part. The aleurone layer accumulates lipids, whereas starchy endosperm mainly accumulates starch. During the ripening stage, the starch accumulation rate is known to be asynchronous, depending on the position of the starchy endosperm. Different physiological and molecular mechanisms are hypothesized to underlie the qualitative and quantitative differences in storage products among developing rice endosperm tissues.
Results
Target cells in aleurone layers and starchy endosperm were isolated by laser microdissection (LM), and RNAs were extracted from each endosperm tissue in the early storage phase. Genes important for carbohydrate metabolism in developing endosperm were analyzed using qRT-PCR, and some of the genes showed specific localization in either tissue of the endosperm. Aleurone layer-specific gene expression of a sucrose transporter, OsSUT1, suggested that the gene functions in sucrose uptake into aleurone cells. The expression levels of ADP-glucose pyrophosphorylase (AGPL2 and AGPS2b) in each endosperm tissue spatially corresponded to the distribution of starch granules differentially observed among endosperm tissues. By contrast, expressions of genes for sucrose cleavage—hexokinase, UDP-glucose pyrophosphorylase, and phosphoglucomutase—were observed in all endosperm tissues tested. Aleurone cells predominantly expressed mRNAs for the TCA cycle and oxidative phosphorylation. This finding was supported by the presence of oxygen (8 % concentration) and large numbers of mitochondria in the aleurone layers. In contrast, oxygen was absent and only a few mitochondria were observed in the starchy endosperm. Genes for carbon fixation and the GS/GOGAT cycle were expressed highly in aleurone cells compared to starchy endosperm.
Conclusions
The transcript level of AGPL2 and AGPS2b encoding ADP-glucose pyrophosphorylase appears to regulate the asynchronous development of starch granules in developing caryopses. Aleurone cells appear to generate, at least partially, ATP via aerobic respiration as observed from specific expression of identified genes and large numbers of mitochondria. The LM-based expression analysis and physiological experiments provide insight into the molecular basis of the spatial and nutritional differences between rice aleurone cells and starchy endosperm cells.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-015-0057-2) contains supplementary material, which is available to authorized users.
Keywords: Aleurone cells, Early storage phase, Laser microdissection, Oryza sativa L, Rice, Starchy endosperm cells
Background
Endosperm accumulates large amounts of nutrients, including starch, proteins, lipids, and minerals, during the ripening stage. Rice (Oryza sativa L.) is the staple food of nearly half of the world’s population (Carriger and Vallee 2007). Rice endosperm is used not only as a source of carbohydrate (mainly starch) energy in the form of steamed rice, but also as a source of oils.
Rice endosperm consists of aleurone cells and starchy endosperm. Lipid is accumulated in aleurone cells, whereas starch is accumulated in starchy endosperm, starting at 5 days after flowering (DAF) (Hoshikawa 1967). Temporal changes in expression of genes for carbohydrate-metabolizing enzymes are closely associated with seed development and sugar status. At the pre-storage phase in rice, the ratio of hexose to sucrose is high, but it starts decreasing with the onset of the storage phase (Ishimaru et al. 2005). Sucrose is the dominant sugar transported into endosperm in the storage phase. Concomitantly, genes important in starch accumulation begin to be expressed at 5 DAF with the onset of starch accumulation. Sucrose is apoplastically unloaded into endosperm from maternal tissues by sucrose transporters. The rice sucrose transporter gene family comprises five genes, OsSUT1-5 (Aoki et al. 2003). Among these five OsSUT genes, OsSUT1 is expressed after 5 DAF (Hirose et al. 1997; Hirose et al. 2002) in aleurone cells in developing endosperm (Furbank et al. 2001; Ishimaru et al. 2007) and plays a critical role in starch accumulation in endosperm (Scofield et al. 2002). After uptake by a sucrose transporter, sucrose is metabolized by sucrose-cleavage enzymes including cell wall invertase and sucrose synthase. These enzymes are crucial for development, growth, and carbon partitioning to sink organs in plants (Sturm and Tang 1999). In rice, eight genes encoding cell wall invertase have been identified, with only OsCIN2 expressed in developing endosperm (Cho et al. 2005). Functional analysis of a rice grain incomplete filling 1 (GIF1) mutant has revealed that the cause is a single mutation in OsCIN2 (Wang et al. 2008). For sucrose synthase, six genes have been identified and spatio-temporal expression in organs has been well characterized. SUS3 and SUS4 are expressed predominantly in developing grains, indicating potential roles in carbon allocation in filling grains (Hirose et al. 2008). Hexokinase (HXK), UDP-glucose pyrophosphorylase, and phosphoglucomutase (PGM) act at an intermediate metabolic step between sucrose cleavage and starch biosynthesis. Ten HXK genes have been cloned, of which five, HXK2, HXK4, HXK5, HXK6, and HXK8, are expressed at a higher level in endosperm than in pericarp, suggesting their role in endosperm (Cho et al. 2006). Two isoforms of UDP-glucose pyrophosphorylase, OsUgp1 and OsUgp2, have been cloned in rice (Chen et al. 2007). Inactivation of OsUgp1 causes grain chalkiness in addition to genic male sterility (Koh et al. 1999; Woo et al. 2008). All of the genes associated with starch biosynthesis, including ADP-glucose pyrophosphorylase (Ohdan et al. 2005), plastid translocator (Toyota et al. 2006), starch synthase (Hirose and Terao 2004), branching enzyme, starch debranching enzyme, phosphorylase, and disproportionating enzyme (Ohdan et al. 2005) have been cloned, and some of them are expressed concomitantly with the onset of endosperm starch accumulation at 5 DAF (Hirose and Terao 2004; Ohdan et al. 2005; Toyota et al. 2006). Genetic analyses using mutants and gene manipulation of starch biosynthesis-related genes have revealed the critical role(s) of some genes in grain phenotypes and starch properties in rice endosperm (Lee et al. 2007; Fujita et al. 2006; Fujita 2014; Umemoto et al. 2004; Fujita et al. 2007; Ryoo et al. 2007; Itoh et al. 2003; Satoh et al. 2003; Satoh et al. 2008; Nishi et al. 2001).
Histological studies have revealed the time course of development of storage product in rice developing endosperm. Aleurone cells begin their differentiation at 4–5 DAF in the outermost endosperm, whereas active starch accumulation starts in the center of the endosperm at 5 DAF (Hoshikawa 1968; Ishimaru et al. 2003). Starch accumulation proceeds asynchronously depending on the region. During the early storage phase, starch rapidly accumulates around the center of the endosperm, whereas in the peripheral starchy endosperm, starch accumulation proceeds at a slower rate until the late storage phase (Hoshikawa 1968). Nitrogen and mineral contents are higher in peripheral endosperm layers corresponding to aleurone cells (Itani et al. 2002). Thus, storage products are quantitatively and qualitatively different depending on their position in the rice endosperm. In barley and maize, histochemical studies have revealed that the oxygen gradient in the endosperm tissues is associated with energy status and the accumulation of storage products. In barley kernels, oxygen-rich regions in the lateral and peripheral endosperm begin starch accumulation first in endosperm tissues under high-ATP conditions, whereas the hypoxic region in the inner endosperm accumulates starch at the later stage (Rolletschek et al. 2004). In developing maize kernels, the oil-storing embryo is in a high-O2 state with high levels of metabolites of glycolytic intermediates and the mitochondrial TCA cycle and with some pools of amino acids (Rolletschek et al. 2005). Thus, in maize and barley, oxygen status differs among endosperm tissues and is closely linked with the accumulation of storage products. However, the quantification of mRNAs of carbohydrate-metabolizing enzymes, described above as important genes for starch accumulation in rice endosperm, was performed using RNAs extracted from whole developing caryopses. No attempt has yet been made to elucidate the asynchronous development of starch among rice endosperm tissues at molecular level. In addition, the oxygen gradient in the developing endosperm is not yet known in rice.
To advance our understanding at the molecular level of positional differences in storage products (lipid and starch) and the starch-accumulating rate in developing rice endosperm, expression analysis by dissection of targeted endosperm tissues is desirable. However, manual dissection of specific tissues is difficult, because aleurone cells and starchy endosperm are structurally connected and very soft in the early storage phase. Laser microdissection (LM) is a powerful tool for isolating targeted individual cells from heterogeneous tissue viewed under a microscope, using an intense laser beam (Emmert-Buck et al. 1996). With LM, we previously succeeded in developing a method for obtaining high-quality RNA from developing rice endosperm, facilitating precise expression analysis (Ishimaru et al. 2007).
In the present study, LM was applied to dissect endosperm tissues at the early storage phase, 7 DAF, when the differentiation of aleurone layers and starchy endosperm is already distinct (Ishimaru et al. 2003) and the degree of starch accumulation varies with endosperm region (Hoshikawa 1968). Using RNAs extracted from each endosperm tissue, qRT-PCR analysis was performed to quantify the expression levels of genes for carbohydrate-metabolizing enzymes. In addition, oxygen concentrations were measured in the developing rice endosperm to determine whether O2 gradients are coupled with different metabolic pathways among endosperm tissues.
Results
Microscopic observation of starch granules and lipids in the endosperm
Under stereomicroscopic observation, the endosperm showed a uniform milky-white color (Fig. 1a), but morphology was qualitatively and quantitatively different among tissues observed with higher magnification. At 7 DAF, the starchy endosperm in the central region (SEC) accumulated much more starch than the starchy endosperm in the lateral regions (SEL) (Fig. 1b). In mature grain, iodine staining was lighter in SEL than in SEC (Fig. 1g). The surrounding outermost cell layer(s) were not stained with iodine solution (Fig. 1c, d, e, g). These cell layer(s) were identified as aleurone cells that were stained with Sudan IV with the accumulation of lipids at maturity (Fig. 1f, h). We defined the dorsal side of aleurone cells as AL (Fig. 1d).
Fig. 1.
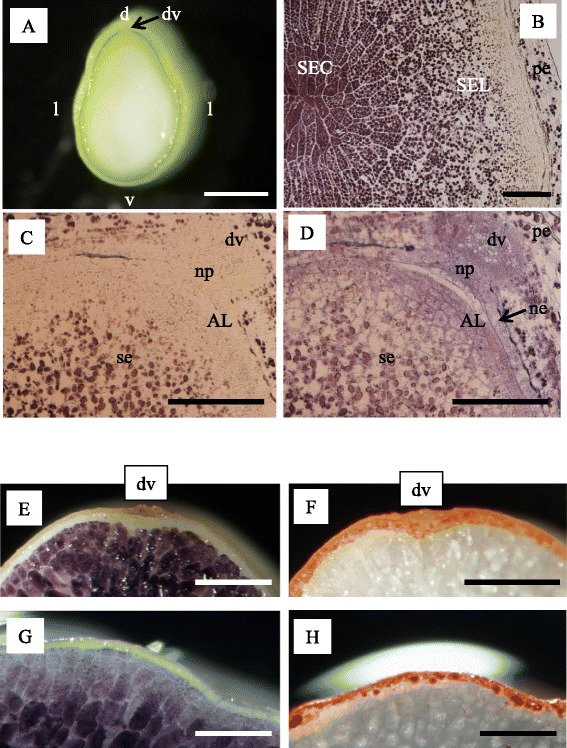
Observation of endosperm cells at 7 DAF (a–d) and in mature grain (e–h). a Median transversal section. b Microscopic observation of starchy endosperm at the center (SEC) and lateral side (SEL) stained with iodine. c, d Microscopic observation of endosperm at the dorsal side stained with iodine (c) and post-stained with toluindine blue-O (d). Matured grain stained with iodine (e, g) and Sudan IV (f, h). AL alurone cells, d dorsal side, dv dorsal vascular bundle, l lateral side, ne nucellar epidermis, np nucellar projection, pe pericarp, v ventral side. Bar: 1 mm (a), 200 μm (b–d), and 500 μm (e–h)
Transmission electron microscopic observation of endosperm cells
The development of organelles in endosperm cells at 7DAF was observed by transmission electron microscopy (TEM). Typical differences were observed in mitochondria in AL and in starch granules in endosperm cells (Fig. 2a and b). The mitochondria in the cells of developing endosperm were quantified along a dorso-ventral axis (the lateral regions of the endosperm are not included in this axis). In the first and second cell layers of the AL, approximately 25 mitochondria were observed per image (310.2 μm2; Fig. 2a, c), but mitochondrial density decreased by half in the third cell layer of the AL (Fig. 2c) and dropped to one fourth in the fifth cell layer of the starchy endosperm compared with the outermost AL cells (Fig. 2b, c). A similar number of mitochondria were observed below cell layers 5 to 7 (Fig. 2).
Fig. 2.
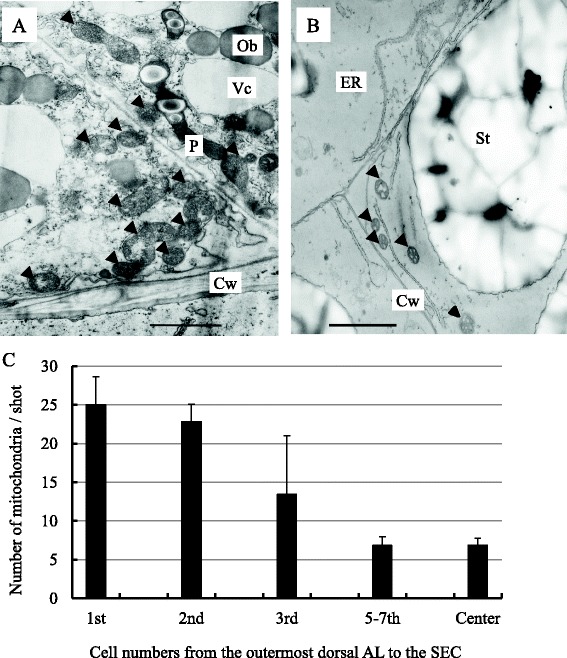
TEM observation of the outermost aleurone cell (a) and central starchy endosperm (b) and number of mitochondria along a dorso-ventral axis (c). Mt mitochondria, St starch, ER endoplasmic reticulum, Vc vacuole, Cw cell wall, Ob oil body, P proplastid. Bar, 2 μm
Oxygen concentration of developing endosperm along a dorso-ventral axis
In view of the differential distribution of mitochondria in the endosperm cell layers, oxygen concentration profiles along the dorso-ventral axis (SEL is not located on the x axis in Fig. 3) in developing rice caryopses at 7 DAF were investigated using a Clark-type O2 microelectrode according to the method of Shimamura et al. (2010). We found that the oxygen concentration decreased steeply as the microelectrode was being inserted deeper into the starchy endosperm. In particular, 8 % oxygen was observed at 200 μm from the surface of the caryopsis (the pericarp), a region corresponding to the dorsal side of AL (Fig. 3). When the electrode was inserted to a depth greater than 300 μm, where the microelectrode had already passed through the AL region and reached the inner starchy endosperm, oxygen concentration was lower than 2 % (Fig. 3). At 1200 μm, a region corresponding to the SEC, oxygen was absent.
Fig. 3.
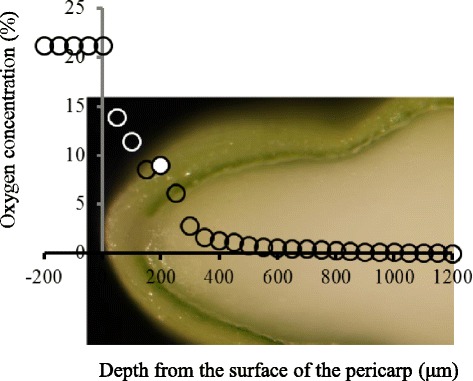
Oxygen concentration (%) profile along a dorso-ventral axis of a developing caryopsis at 7 DAF. The x-axis indicates the approximate position where the microelectrode was inserted. The white enclosed circle at 200-μm depth corresponds to the position of dorsal aleurone cells. An oxygen concentration profile was obtained from three kernels and values at each depth were averaged
LM for endosperm cells at 7 DAF and identity confirmation of dissected tissues by qRT-PCR
An overview of developing endosperm at 7 DAF is shown in Fig. 4a and b. AL (Fig. 4c, d), SEC (Fig. 4e), and SEL (Fig. 4e, f) were isolated by LM. We note that the aleurone cell layer at the outermost endosperm cells in the lateral regions was not isolated during dissection of the SEL (Fig. 4e, f). High-quality RNA, with RNA integrity number (RIN) > 7.0, was obtained from dissected tissues (Additional file 1: Table S1). The identities of dissected tissues were confirmed by qRT-PCR using primers for tissue-specific mRNAs for oleosin, 16 kDa isoform R16 (NCBI: AF022148), a structural protein found in oil bodies involved with lipid accumulation, and starch debranching enzyme (SDBE, Nakamura et al. 1996; NCBI: D50602), which specifically degrades amylopectin (Table 1). Expression values from quantitative RT-PCR were normalized to the transcript level of 18S rRNA (Kim et al. 2003; Additional file 2: Figure S1). The mRNAs for oleosin, 16 kDa isoform and SDBE were supposed to be markers specific for AL and starchy endosperm, respectively, because of the specific localization of starch granules and lipids in the endosperm (Fig. 1). Transcripts of oleosin, 16 kDa isoform R16, were detectable only in AL, whereas those of SDBE were at almost negligible levels in AL (Additional file 3: Figure S2A). The transcript level of SDBE was higher in the SEC than in the SEL (Additional file 3: Figure S2B) consistent with the gradients shown by iodine staining (Fig. 1b). These results confirmed the precise dissection of targeted endosperm tissues.
Fig. 4.
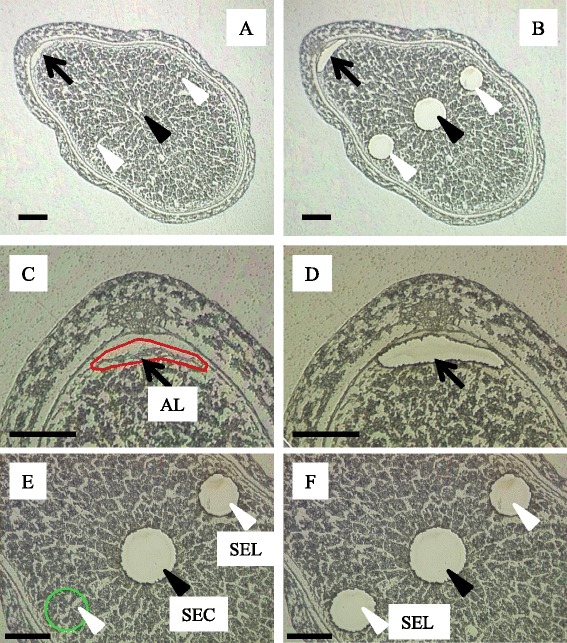
Microdissection of endosperm tissues. a Median transverse section. b Microdissection of aleurone cells from the dorsal side (AL; black arrow, magnified in c and d), starchy endosperm from the center region (SEC; black arrowhead, magnified in e and f) and lateral regions (SEL; white arrowheads, magnified in e and f). Bar: 200 μm
Table 1.
Internal control and marker genes for aleurone cells and starchy endosperm
| Category | NCBI accession number | Description | Reference |
|---|---|---|---|
| Internal control | X00755 | 18S rRNA | Kim et al. 2003 |
| Marker for aleurone cells | AF022148 | 16 kDa oleosin isoform R16 | NCBIa; Medina and Quatrano, unpublished. |
| Marker for starchy endosperm | D50602 | Starch debranching enzyme | Pullulanase; Nakamura et al. 1996, OsPUL; Ohdan et al. 2005 |
aDirect submission to The National Center for Biotechnology Information (NCBI)
qRT-PCR for genes associated with sucrose transport, sucrose cleavage, and starch biosynthesis
Transcription levels of genes associated with sucrose transport (Hirose et al. 1997), sucrose cleavage (cell wall invertase; Cho et al. 2005 and sucrose synthase; Hirose et al. 2008), hexokinase (Cho et al. 2006), UDP-glucose pyrophosphorylase (Chen et al. 2007), phosphoglucomutase (Akiyama et al. unpublished), and plastidis translocator (Toyota et al. 2006) and starch biosynthesis (Hirose and Terao 2004; Ohdan et al. 2005) during rice grain filling were evaluated. The selected genes are considered to play dominant roles in the developing endosperm among gene families with high expression or critical roles in grain phenotype and starch properties, according to previous genetic studies (see review by Fujita 2014, Table 2, and Background).
Table 2.
Information on the genes related to sucrose transport, sucrose cleavage, intermediate metabolic steps and starch biosynthesis
| Category | Accession No. | Description | Reference | Major changes in grain appearance and starch properties with the mutant and genetic manipulation of gene |
|---|---|---|---|---|
| Sucrose transport and cleavage | D87819 | Sucrose transporter (SUT1) | OsSUT1; Hirose et al. 1997 | Impaired grain filling (Scofield et al. 2002) |
| AK072276 | Cell wall invertase 2 | OsCIN2; Cho et al. 2005 | Chalky phenotype with abnormal amyloplast (gif1; Wang et al. 2008) | |
| AK100306 | Sucrose synthase 3 | SUS3; Hirose et al. 2008 | - | |
| AK102158 | Sucrose synthase 4 | SUS4; Hirose et al. 2008 | - | |
| Metabolic step between sucrose cleavage and starch biosynthesis | DQ116384 | Hexokinase 2 | OsHXK2; Cho et al. 2006 | - |
| DQ116386 | Hexokinase 4 | OsHXK4; Cho et al. 2006 | - | |
| DQ116387 | Hexokinase 5 | OsHXK5; Cho et al. 2006 | - | |
| DQ116388 | Hexokinase 6 | OsHXK6; Cho et al. 2006 | - | |
| DQ116390 | Hexokinase 8 | OsHXK8; Cho et al. 2006 | - | |
| AB062606 | UDP-glucose pyrophosphorylase | UGPase; Abe et al. 2002, OsUgp1; Chen et al. 2007, UGPase1; Woo et al. 2008 | Chalky phenotype (Koh et al. 1999) | |
| AF455812 | Phosphoglucomutase | NCBIa; Akiyama, unpublished. | - | |
| Starch biosynthesis | U66041 | ADP-glucose pyrophosphorylase large subunit | OsAGPL2; Ohdan et al. 2005; Lee et al. 2007 | shrunken phenotype (Lee et al. 2007) |
| AK103906 | ADP-glucose pyrophosphorylase small subunit | OsAGPS2b; Ohdan et al. 2005; Lee et al. 2007 | shrunken phenotype (Lee et al. 2007) | |
| AK107368 | ADP-glucose transporter | OsBT1-1; Toyota et al. 2006 | brittle phenotype (maize; Shannon et al. 1998, barley; Patron et al. 2004) | |
| D16202 | Soluble starch synthase 1 | SSS; Baba et al. 1993, SSI; Hirose and Terao 2004, OsSSI; Ohdan et al. 2005 | Altered fine structure of amylopectin (Fujita et al. 2006) | |
| AF419099 | Soluble starch synthase II-3 | SSII-3; Hirose and Terao 2004, SSIIa; Ohdan et al. 2005 | Altered fine structure of amylopectin (alk; Umemoto et al. 2004) | |
| AY100469 | Soluble starch synthase III-2 | SSIII-2; Hirose and Terao 2004, OsSSIII-2; Dian et al. 2005, OsSSIIIa; Ohdan et al. 2005 | White-cored chalky phenotype and altered fine structure of amylopectin (Fujita et al. 2007, flo5; Ryoo et al. 2007) | |
| X62134 | Granule-bound starch synthase I | Okagaki 1992, GBSSI; Hirose and Terao 2004, OsGBSSI; Ohdan et al. 2005 | waxy phenotype with the absence of amylose (Itoh et al. 2003) | |
| D11082 | Starch branching enzyme I | RBEI; Mizuno et al. 1992, OsBE1; Ohdan et al. 2005 | Altered fine structure of amylopectin (sbeI; Satoh et al. 2003) | |
| D16201 | Starch branching enzyme IIb | RBEIII; Mizuno et al. 1993, OsBEIIb; Ohdan et al. 2005 | Chalky phenotype and altered fine structure of amylopectin (amylose-extender; Nishi et al. 2001) | |
| AB093426 | Starch debranching enzyme: Isoamylase I | OsISA1; Ohdan et al. 2005 | sugary phenotype (sugary-1; Kubo et al. 1999a) | |
| AK063766 | Plastidial phosphorylase | OsPHOL; Ohdan et al. 2005 | shrunken to pseudonormal phenotypes (pho1; Satoh et al. 2008), protein phosphorylation in amyloplast (wheat; Tetlow et al. 2004) |
aDirect submission to The National Center for Biotechnology Information (NCBI)
The expression of OsSUT1 was specific to aleurone cells and undetectable in the SEL and SEC. The expression levels were contrasting among the genes for sucrose-cleavage enzymes. The highest expression levels for OsCIN2, OsSUS3, and OsSUS4 were observed in AL, SEL, and SEC, respectively (Fig. 5).
Fig. 5.
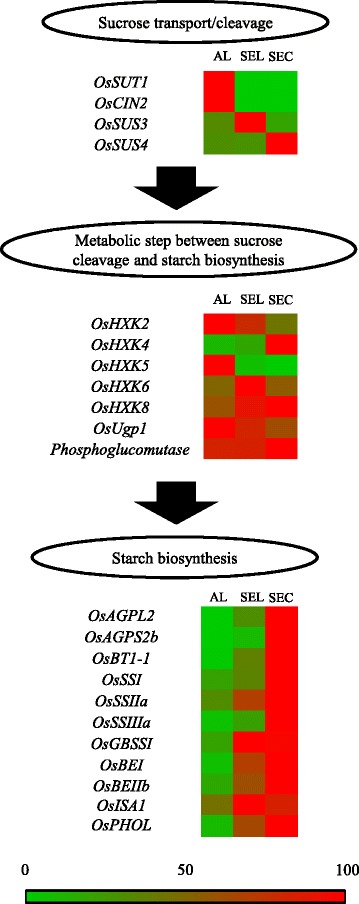
qRT-PCR analysis of sucrose transport, sucrose cleavage, intermediate metabolic steps and starch biosynthesis. Values are shown in a sequential color chart from 0 (green) to 100 (red). The tissue with the highest value among endosperm tissues was adjusted to 100. The values are means of three biological replicates. The accession numbers and primer pairs for each gene are given in Table 2
A major role of hexokinase (OsHXK) genes in rice developing endosperm has not yet been clarified by genetic approaches, but the genes OsHXK2, OsHXK4, OsHXK5, OsHXK6, and OsHXK8 were selected in view of their preferential expression in developing endosperm, based on the report of Cho et al. (2006). OsHXK4 and OsHXK5 were expressed predominantly in SEC and AL, respectively. The expression of OsHXK2 and OsHXK6 was observed in all tissues, but preferentially in AL and SEL, respectively. The expression of OsHXK8 was higher in SEL and SEC compared to AL. Expression of OsUgp1 and phosphoglucomutase was observed in all tissues.
The expression of genes for starch biosynthesis was variable. Expression was at low level (with values below 25) in AL for all genes except OsSSIIa and OsISA1. The expression of OsAGPL2 and OsAGPS2b, OsBT1-1, OsSSI, OsSSIIIa, and OsPHOL was preferential in SEC. The expression of genes for OsSSIIa, OsBEI, and OsBEIIb was relatively high (with values over 60) in the SEL and highest in the SEC. The expression of genes for OsGBSSI and OsISA1 was highest in SEL among endosperm tissues tested. We also investigated the expression of disproportionating enzyme (OsDPE1; Ohdan et al. 2005), which plays a critical role in building the amylopectin structure in Chlamydomonas reinhardtii (Colleoni et al. 1999). The expression of OsDPE1 was undetectable in any tissues at 7DAF (data not shown), possibly because the transcript level abruptly decreases after 5DAF in the developing caryopsis (Ohdan et al. 2005).
qRT-PCR of genes associated with CO2 fixation, the TCA cycle, oxidative phosphorylation, and the GS/GOGAT cycle
The presence of oxygen (8 %) in the AL (Fig. 3), suggested the high expression of genes associated with aerobic respiration. In addition to the expression of genes for TCA cycle and oxidative phosphorylation, the expression of genes associated with CO2 fixation and the GS/GOGAT cycle, a metabolic step is close to the TCA cycle and oxidative phosphorylation, was evaluated (Table 3; Fig. 6). Some of the selected genes have not yet been characterized, but they appear in NCBI (http://www.ncbi.nlm.nih.gov/nuccore/) and RAP-DB (http://rapdb.dna.affrc.go.jp/) as our target genes based on their sequence similarity to genes in other plant species (Table 3).
Table 3.
Information on the genes related to carbon fixation, TCA cycle, oxidative phosphorylation and GS/GOGAT cycle
| Category | Accession No. | Description | Reference |
|---|---|---|---|
| Carbon fixation | AK059261 | Acetyl CoA carboxylase | RAP-DBa; Similar to Acetyl-coenzyme A carboxylase. (Os05t0295300-01) |
| AF271995 | Phosphoenolpyruvate carboxylase 1 | Osppc1; Yamamoto et al. 2014 | |
| TCA cycle | AF302906 | Citrate synthase | NCBIb; Silva et al. Unpublished. |
| AK067183 | Aconitase | RAP-DBa; Similar to Aconitate hydratase, cytoplasmic (Citrate hydro-lyase) (Aconitase). (Os03t0136900-01). Similar to Aconitate hydratase 2, mitochondrial. (Os03t0136900-02) | |
| AF155333 | NADP-isocitrate dehydrogenase | Koyama et al. 1999 | |
| AK100482 | Oxoglutarate dehydrogenase | RAP-DBa; Similar to 2-oxoglutarate dehydrogenase, E1 component. (Os07t0695800-01) | |
| AK103525 | Succinyl-CoA synthetase | RAP-DBa; Similar to Succinyl-CoA synthetase, beta chain. (Os02t0621700-01) | |
| AB017428 | Succinate dehydrogenase | SDHB; Kubo et al. 1999b | |
| AF444195 | Malate dehydrogenase 1 | MDH; Lin et al. 2003 | |
| AK073698 | Malate dehydrogenase 2 | RAP-DBa; Similar to Malate dehydrogenase. (Os05t0574400-01) | |
| Oxidative phosphorylation | AK058713 | NADH-ubiquinone oxidoreductase | RAP-DBa; Similar to NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial precursor. (Os03t0713400-01); Similar to NADH-ubiquinone oxidoreductase 75 kDa subunit. (Os03t0713400-02); Similar to NADH-ubiquinone oxidoreductase 75 kDa subunit. (Os03t0713400-03) |
| AK243655 | NADH-ubiquinone oxidoreductase subunit PSST | RAP-DBa; NADH-ubiquinone oxidoreductase subunit PSST (Fragment). (Os05t0533700-01) | |
| AK119716 | Cytochrome b-c1 complex subunit 8 | RAP-DBa; Similar to Cytochrome b-c1 complex subunit 8. (Os06t0175900-01) | |
| GS/GOGAT cycle | Y12594 | Ferredoxin-dependent glutamate synthase (Fd-GOGAT) | OsGog1; Mattana et al. Unpublishedb. |
| X14245 | Cytosolic glutamine synthetase 1;1 | GS1; Sakamoto et al. 1989, GS1;1; Tabuchi et al. 2005 | |
| X14244 | Cytosolic glutamine synthetase 1;2 | GSr; Sakamoto et al. 1989, GS1;2; Tabuchi et al. 2005 | |
| AB008845 | NADH dependent Glutamate Synthase | Goto et al. 1998 | |
| AY332470 | Glutamate dehydrogenase | OsGDH1.2; Qiu et al. 2009 |
aThe Rice Annotation Project Database
bDirect submission to The National Center for Biotechnology Information (NCBI)
Fig. 6.
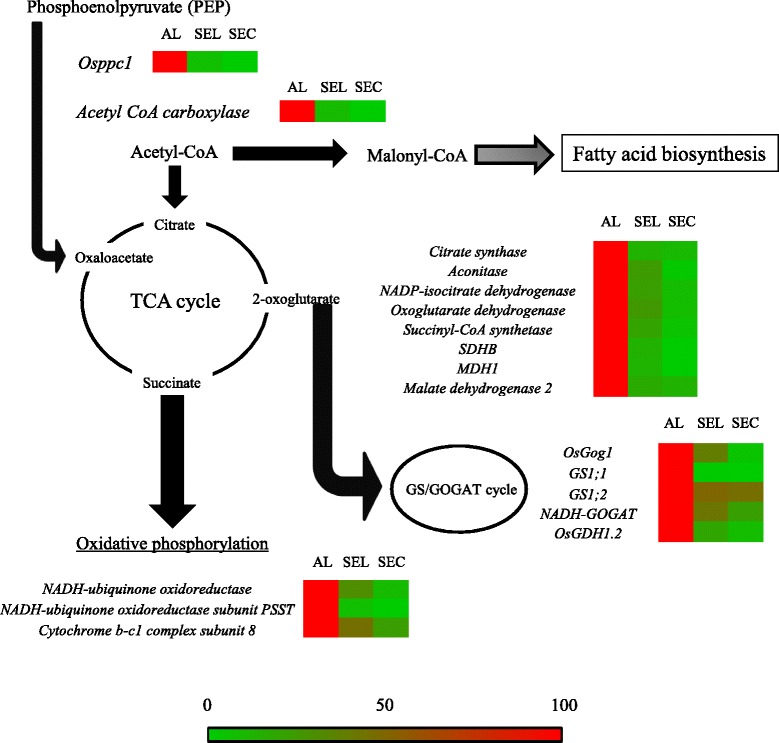
qRT-PCR analysis for carbon fixation, TCA cycle, oxidative phosphorylation, and GS/GOGAT cycle. Succinate dehydrogenase is also classified as a gene for oxidative phosphorylation. The sequential color chart refers to Fig. 4. The accession numbers and primer pairs for each gene are given in Table 3
All the genes analyzed showed highest expression in AL, and their expression levels decreased in the order SEL > SEC (Fig. 6). Some of the genes were localized predominantly in the AL (Osppc1, Acetyl-CoA carboxylase, genes for the TCA cycle, NADH-ubiquinone oxidoreductase subunit PSST, GS1;1, OsGDH1.2).
Discussion
Transcripts associated with metabolic steps from sucrose transport to starch biosynthesis were differentially distributed in developing endosperm
Genetic analyses using mutants and gene manipulation have revealed the critical role(s) of genes for carbohydrate-metabolizing enzymes in kernel phenotypes and starch properties (Table 2). In this study, LM was applied to the AL, SEL and SEC (Fig. 4) to quantify the distribution of transcripts at metabolic steps from sucrose transport to starch biosynthesis, relative to their spatial distribution in starch granules among tissues in the early storage phase (Fig. 1).
The transcript of sucrose transporter 1 (OsSUT1; Hirose et al. 1997) was specific to AL, as we reported previously for AL and SEC (Ishimaru et al. 2007). This study showed negligible expression in SEL (Fig. 5). An anti-sense transformant of OsSUT1 showed impaired grain filling (Scofield et al. 2002). The specific localization of OsSUT1 in AL (Fig. 5) suggests the critical function of aleurone cells for uptake of sucrose into developing endosperm tissues for starch accumulation. Our LM-based expression analysis revealed the specific localization of OsCIN2 in AL among endosperm tissues as that of OsSUT1 (Fig. 5). A mutation of OsCIN2 (gif1) causes slower grain filling and chalky phenotype of the kernel with aberrant amyloplast formation, indicating that hexoses catalyzed by OsCIN2 function as an important carbon energy source for grain development in rice (Wang et al. 2008). The specific distribution of OsCIN2 in AL (Fig. 5) was spatially inconsistent with starch accumulation in the starchy endosperm, such as in SEL and SEC. OsCIN2 in AL is expected to contribute to the partitioning of hexoses to the inner endosperm tissues for starch accumulation in SEL and SEC. The organ expression analysis of OsSUS3 and OsSUS4 revealed their predominant localization in the developing caryopsis, especially in the endosperm, after the onset of starch accumulation at 5 DAF (Hirose et al. 2008). Our LM-based expression analysis found OsSUS3 and OsSUS4 to be localized mainly in the SEL and SEC, respectively (Fig. 5). The expression profile of the sucrose-cleavage genes OsCIN2, OsSUS3, and OsSUS4 was spatially complemented in the developing endosperm. The cleavage of sucrose in the developing endosperm is necessary for generating a sucrose gradient to maintain sink strength (Sturm and Tang 1999). The results suggest that OsCIN2, OsSUS3, and OsSUS4 contribute to sink strength by cleaving sucrose in different locations in developing endosperm tissues. With respect to the expression profile for hexokinase, OsHXK4 and OsHXK5 showed predominant expression in SEC and AL, respectively. For OsHXK2, a gradient in expression level was observed from AL to SEC in descending order. OsHXK6 and OsHXK8 showed highest expression level in SEL and SEC, respectively, and relatively high expression was observed remaining two tissues. Transcripts of OsUgp1 and phosphoglucomutase were highest in AL and SEC, respectively, and relatively high expression was observed in the remaining two tissues. Thus, the expression of each gene appeared to be functional in a different space in hexokinase. OsUgp1 and phosphoglucomutase are assumed to generate metabolites for subsequent starch biosynthesis in all three regions: AL, SEL, and SEC.
With respect to genes for starch biosynthesis, expression was low in AL, and the highest values, except for OsGBSSI and OsISA1, were observed in SEC (Fig. 5). Expression of OsAGPL2, OsAGPS2b, OsBT1-1, OsSSI, and SSIIIa was low in SEL, whereas expression of OsSSIIa, OsGBSSI, OsBEI, OsBEIIb, and OsPHOL was relatively high (with values over 60) in SEL. T-DNA insertion into either OsAGPL2 or OsAGPS2b resulted in a shrunken phenotype of rice kernels with impaired grain filling (Lee et al. 2007). Mutant plants of brittle-1 in maize (Shannon et al. 1998) and lys5 in barley (Patron et al. 2004) show drastically decreased kernel dry weight, owing to the absence of a plastidial ADP-glucose transporter. The gradient of transcript abundance for OsAGPL2, OsAGPS2b, and OsBT1-1 was consistent with the gradient of starch accumulation among AL, SEL, and SEC (Fig. 1), suggesting the in vivo regulation of starch accumulation by these genes at the transcriptional level. For starch synthase (both soluble and granule-bound type), starch branching enzymes, and starch debranching enzyme, greater attention has been paid to the effects of genes on starch properties such as amylopectin structure and amylose formation. Relatively high levels of expression of OsSSIIa, OsGBSSI, OsBEI, OsBEIIb, OsISA1, and OsPHOL in SEL suggest that these genes are responsible for the production of amylopectin and amylose in SEL at the transcriptional level. The subcellular location of OsAGPL2 and OsAGPS2b is cytosolic (Sikka et al. 2001), whereas that of OsBT1-1, a gene for a starch synthase branching enzyme, and starch debranching enzyme is plastidial. In addition, the metabolic step of ADP-glucose pyrophosphorylase is located upstream from that of starch synthase, starch branching enzyme, and starch debranching enzyme. In SEL, the substrate of ADP-glucose may be deficient in plastids, owing to the very low level of cytosolic OsAGPL2 and OsAGPS2b. Starch biosynthesis in the cereal endosperm is a complex process engaged with many genes (Fig. 5). Overall results of expression of genes associated with metabolic steps from sucrose transport to starch biosynthesis revealed the clear differences in the distribution of transcripts in the developing endosperm. The LM-based expression analysis conducted in this study provided the novel finding that the clear gradient of transcripts of OsAGPL2 and OsAGPS2b may be responsible for the large difference in starch accumulation among AL, SEL, and SEC in the early storage phase.
Aleurone cells are inferred to generate ATP by aerobic respiration during the early storage phase
Aleurone cells do not contain starch, but contain lipids (Fig. 1e and f). Our LM-based expression analysis showed the specific localization of oleosin, 16 kDa isoform R16, in AL (Additional file 3: Figure S2), and the low level of transcripts for starch biosynthesis in AL (Fig. 5). The expression analysis is consistent with the large amount of lipid and the absence of starch in AL (Fig. 1c). In developing maize, oxygen concentration is maintained at the high level in the oil-storing embryo, corresponding to the steady-state levels of glycolytic intermediates and those of the TCA cycle, as well as free amino acids (Rolletschek et al. 2005). In the present study, we investigated the spatial distribution of oxygen (Fig. 3) and transcripts of genes associated with carbon fixation, the TCA cycle, oxidative phosphorylation, and the GS/GOGAT cycle (Fig. 6). Oxygen was detectable in the outermost endosperm cells corresponding to the aleurone layers at 8 % (Fig. 3). All the genes examined in Table 3 were expressed dominantly in AL (Fig. 6), supporting the findings in maize embryo (Rolletschek et al. 2005) with respect to oxygen concentration and metabolites in oil-storage tissue. In the present study, we identified clear gradients in the number of mitochondria from the AL to the SEC in descending order (Fig. 2). Oparka et al. (1981) reported the presence of mitochondria in the aleurone and sub-aleurone layers with TEM observation. The present quantitative investigation showed that the profile of oxygen distribution agrees well with the gradient in numbers of mitochondria in rice endosperm in the early storage phase. Xu et al. (2008) reported that accumulation of proteins associated with the TCA cycle increased during 6–10 DAF (in the early storage phase), based on proteomic analysis of developing rice kernels. Thus, mitochondria localized in the oxygen-rich cells of AL are expected to contribute to ATP generation through aerobic respiration, thereby assisting the initial formation and accumulation of lipids via expression of genes listed in Table 3. In barley, starch accumulation is initiated in the lateral region, where the tissues retain a high level of oxygen supplied by photosynthesis in the surrounding pericarp (Rolleschek et al. 2004). In rice, starch accumulation is initiated in the center of the endosperm (Fig. 1b), where the tissue is most distant from the pericarp and in a condition of hypoxia (Fig. 3). Energy production for starch accumulation may be different between barley and rice, given the differences in oxygen availability in the starch-storing tissue in the early storage phase. SEC is assumed to produce ATP via anaerobic respiration to supply energy for starch development in the absence of oxygen (Fig. 3). Whether gradients in oxygen concentration are coupled with the differences in storage product between endosperm tissues (i.e. lipids in aleurone cells and starch in starchy endosperm) through the different process of energy production is still elusive. Further evidence are required to reveal the relationship between the energy production and biological process of storage product accumulation in the different positions of developing endosperm. Knockout mutants of cytosolic glutamine synthetase 1;1 showed a significant reduction in rice kernel weight (Tabuchi et al. 2005), indicating the involvement of this gene in carbon partitioning through nitrogen metabolism in the AL. The function of cytosolic glutamine synthetase 1;1 in the developing endosperm is still unclear, but there may be physiological linkages between nitrogen metabolism in aleurone cells and starch synthesis in starchy endosperm.
Conclusions
We revealed the expression pattern of carbohydrate-metabolizing genes in the different positions of developing endosperm with an assistance of LM. The expression of OsSUT1 was specific to the AL, and the expression of sucrose-cleavage enzymes such as OsCIN1, OsSUS3, and OsSUS4 was preferential in AL, SEL and SEC, respectively. The gradients of transcript abundance for OsAGPL2 and OsAGPS2b were assumed to be associated with the differential spatial distribution of starch granules among endosperm tissues in the early storage phase. These results in carbohydrate-metabolizing genes suggested the roles of each gene in carbon partitioning and starch synthesis in the different positions of developing endosperm. The presence of oxygen and large number of mitochondria in AL were consistent with the predominant expression of genes involved in TCA cycles and oxidative phosphorylation, inferring the energy production via aerobic respiration at least in part in AL. The LM-based expression analysis conducted in this study expanded molecular and physiological knowledge on the positional differences in starch accumulation and energy production in the developing rice endosperm in the early storage phase.
Methods
Plant materials
Oryza sativa cv. Koshihikari (a Japonica rice variety) was used. Seeds were sown in a nursery box filled with soil, and 4 weeks-old seedlings were transplanted into 0.02 m2 pots. As a basal dressing, 0.5, 2.3, and 2.2 g each of N, P2O5, and K2O was applied, and 0.4 g of nitrogen per pot was applied as a top dressing approximately two weeks before heading. At the booting stage, plants were transferred into a naturally illuminated temperature-controlled chamber. Day (13 h) and night (11 h) air temperatures were maintained at 26 and 20 °C, respectively until maturity. Spikelets were marked on the flowering day. Caryopses at 7 DAF located on the four primary rachis branches counted from the top of the panicles were used in all experiments.
Laser microdissection (LM)
Four developing rice caryopses were collected from one panicle. Three biological replicates (panicles) were prepared from three rice plants. The preparation of specimens for LM followed Ishimaru et al. (2007). Briefly, the developing caryopses were immediately fixed with an ice-cold mixture of 3:1 ethanol: acetic acid, and embedded with 2 % carboxymethylcellulose. Transverse sections (8 μm thickness) were made at the median part of the developing caryopses with a cryomicrotome (Leica, CM1850). Aleurone cells at the dorsal side (AL) and starchy endosperm in the center (SEC) and the lateral regions (SEL) were microdissected with an AS LMD system (Leica Microsystems, Wetzlar, Germany).
RNA extraction, quantification, and quality check
Total RNA was extracted with a Picopure RNA isolation kit (Molecular Devices, Sunnyvale, CA) using DNase I. Quantification of total RNA was determined by the fluorescence based method, using a RiboGreen RNA Quantification kit (Molecular Probes, Eugene, OR). The integrity of RNA from aleurone cells and starchy endosperm was assessed using a 2100 Bioanalyzer (Agilent technologies, Santa Clara, CA). The average RNA integrity number (RIN) with clear rRNA peaks from each tissue exceeded 7.0 for the three biological replications (Additional file 1: Table S1).
cDNA synthesis and quantitative RT-PCR
Quantitative RT-PCR was performed following Ishimaru et al. (2009) with three biological replications. Total RNA (10 ng) was amplified and cDNA was synthesized with a WT-OvationTM RNA Amplification System (NuGEN technologies Inc., San Carlos, CA) according to the manufacturer’s instructions. For quantitative RT-PCR, SYBR Premix Ex Taq (TaKaRa Bio Inc, Shiga, Japan) was used with a real time RT-PCR system (7500 Real Time PCR System, Applied Biosystems, Foster, CA). Gene-specific primer pairs were designed with Primer 3 (version 0.4.0., http://frodo.wi.mit.edu/primer3/) or taken from previous reports (Additional file 4: Table S2). Dissociation curves confirmed the presence of a single amplicon in each PCR. cDNA of each gene was further amplified with Taq polymerase (ExTaq; TaKaRa Bio Inc), and PCR products were sequenced to confirm that the fragments were the targeted gene. For each gene, the highest value in a given tissue was adjusted to 100 after normalization with the transcript level of 18S rRNA (Kim et al. 2003; Additional file 2: Figure S1), and relative values less than 100 were determined for two other tissues.
Oxygen concentration in developing caryopses
Oxygen concentration in developing caryopses was measured following Shimamura et al. (2010). Immediately after the detachment of the developing caryopsis from the glume, the caryopsis was placed in an aluminum block and fixed with dental silicone impression material (Provil novo light, Heraeus Kulzer GmbH, Germany). The base of the rachilla was covered with dental silicone impression material to prevent an inflow of external air after detachment. The impression material hardened within 2 min. A Clark-type O2 microelectrode (OX-25, Unisence A/S, Denmark) with a guard cathode and a tip diameter of 25 μm was inserted into a developing caryopsis at 50-μm intervals along a dorso-ventral axis. The microelectrode was connected to a pA meter (PA2000, Unisense A/S) and output was logged at 5-s intervals on a computer using an analog-to digital converter (ADC-16, Pico Technology, UK). The electrode was calibrated in air before measurement and in O2-free N2. The experiment was conducted at a temperature of 25 °C and 12 μmol m−2 s−1 photon flux density. Three developing caryopses sampled from different panicles were used. O2 concentrations obtained from three developing caryopses were averaged.
Microscopic observation
Stereo microscope
Median transverse sections (1.0–1.5 mm thickness) of developing caryopses and matured grain were manually cut with a sharp razor. Sectioned mature kernels were immersed into a solution of 2 % KI and 0.4 % I2 or 2 % Sudan IV (w/v) in 70 % ethanol for staining starch or lipid, respectively. After staining, specimens were viewed under a stereomicroscope (SZX12, Olympus, Japan), and immediately photographed.
Light microscope
Developing caryopses were immersed in FAA (formalin: acetic acid: 70 % ethanol = 1: 1: 18), dehydrated in an ethanol series, and embedded in Technovit 7100 (Heraeus Kulzer GmbH, Hanau, Germany). Sections 3 μm thick were cut with a microtome (HM335E, Leica Microsystems, Germany). Sections were stained with a solution of 2 % KI and 0.4 % I2 to observe starch granules and the same specimens were then stained again with 0.1 % toluidine blue-O to observe median transverse endosperm.
Transmission electron microscope (TEM)
Developing caryopses at 7 DAF were hand-cut into 1.0 mm-thick sections at the median with a sharp razor and immediately fixed with ice-cold 4.0 % paraformaldehyde and 2.0 % glutaraldehyde for 3 h. Specimens were fixed again overnight at 4 °C. After washing with 100 mM phosphate buffer (pH 7.2), the specimens were post-fixed with 1 % osmium tetroxide overnight at 4 °C. They were then washed with distilled water, dehydrated in an ethanol series, and embedded in Spurr’s resin (Spurr Low Viscosity Embedding kit, Polyscience Inc., Warrington, PA). Sections 200 nm thick were cut with glass knives with an ultramicrotome (MT2-B, Sorvall, Newtown, CT) at the median part of the specimen. Sections were stained with TI blue (Nissin EM, Tokyo, Japan) for 2 h and with lead citrate for 7 min, and viewed under an H-7100 transmission electron microscope (Hitachi, Tokyo, Japan) at 75 kV. Mitochondria were counted from the images at ×3500 magnification on 76 × 50 mm films (310.2 μm2). Mean values were calculated with 13–29 independent images (cells) from two biological replications.
Acknowledgements
T. I. gratefully acknowledges Prof. N. Tsutsumi (The University of Tokyo) for technical advice on the measurement of mitochondria. T. I. also thanks Dr. Taniguchi (NICS), Dr. Ishikawa (NICS), Ms. Ito, and Ms. Hiroki for technical assistance. This work was supported by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No.20780013 to T. I.).
Additional files
RNA integrity number (RIN) used for qRT-PCR analysis.
qRT-PCR for 18S rRNA. The gene accession numbers and primer pairs are shown in Table 1. Values are the means of three biological replications. The value in each tissue was used for the normalization.
qRT-PCR for 16 kDa oleosin (A) and starch debranching enzyme (B; SDBE). The gene accession numbers and primer pairs are shown in Table 1. Values are the means of three biological replications.
Gene-specific primers used in this study.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TI, TM, MN, and MK designed the experiments. TI, MI, SH, and SS conducted the experiments. TI, SS, TM, NKN, MN, and MK wrote the manuscript. All authors read and approved the submitted version of manuscript.
References
- Abe T, Niiyama H, Sasahara T. Cloning of cDNA for UDP-glucose pyrophosphorylase and the expression of mRNA in rice endosperm. Theor Appl Genet. 2002;105:216–221. doi: 10.1007/s00122-002-0927-z. [DOI] [PubMed] [Google Scholar]
- Aoki N, Hirose T, Scofield GN, Whitfeld PR, Furbank RT. The sucrose transporter gene family in rice. Plant Cell Physiol. 2003;44:223–232. doi: 10.1093/pcp/pcg030. [DOI] [PubMed] [Google Scholar]
- Baba T, Nishihara M, Mizuno K, Kawasaki T, Shimada H, Kobayashi E, Ohnishi S, Tanaka K, Arai Y. Identification, cDNA cloning, and gene expression of soluble starch synthase in rice (Oryza sativa L.) immature seeds. Plant Physiol. 1993;103:565–573. doi: 10.1104/pp.103.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriger S, Vallee D. More crop per drop. Rice Today. 2007;6:10–13. [Google Scholar]
- Chen R, Zhao X, Shao Z, Zhu L, He G. Multiple isoforms of UDP-glucose pyrophosphorylase in rice. Physiol Planta. 2007;129:725–736. doi: 10.1111/j.1399-3054.2007.00865.x. [DOI] [Google Scholar]
- Cho JI, Lee SK, Ko S, Kim HK, Jun SH, Lee YH, Bhoo SH, Lee KW, An G, Hahn TR, Jeon JS. Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.) Plant Cell Rep. 2005;24:225–236. doi: 10.1007/s00299-004-0910-z. [DOI] [PubMed] [Google Scholar]
- Cho JI, Ryoo N, Ko S, Lee SK, Lee J, Jung KH, Lee YH, Bhoo SH, Winderickx J, An G, Hahn TR, Jeon JS. Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.) Planta. 2006;224:598–611. doi: 10.1007/s00425-006-0251-y. [DOI] [PubMed] [Google Scholar]
- Colleoni C, Dauvillée D, Mouille G, Morell M, Samuel M, Slomiany MC, Liénard L, Wattebled F, d’Hulst C, Ball S. Biochemical characterization of the Chlamydomonas reinhardtii α-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiol. 1999;120:1005–1014. doi: 10.1104/pp.120.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dian W, Jiang H, Wu P. Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot. 2005;56:623–632. doi: 10.1093/jxb/eri065. [DOI] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Fujita N. Starch biosynthesis in rice endosperm. AGri-Biosci Monographs. 2014;4:1–18. doi: 10.5047/agbm.2014.00401.0001. [DOI] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y. Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 2006;140:1070–1084. doi: 10.1104/pp.105.071845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park JH, Jane JL, Miyao A, Hirochika H, Nakamura Y. Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007;144:2009–2023. doi: 10.1104/pp.107.102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT, Scofield GN, Hirose T, Wang XD, Patrick JW, Offler CE. Cellular localization and function of a sucrose transporter OsSUT1 in developing rice grains. Aust J Plant Physiol. 2001;28:1187–1196. [Google Scholar]
- Goto S, Akagawa T, Kojima S, Hayakawa T, Yamaya T. Organization and structure of NADH-dependent glutamate synthase gene from rice plants. Biochim Biophys Acta. 1998;1387:298–308. doi: 10.1016/S0167-4838(98)00142-3. [DOI] [PubMed] [Google Scholar]
- Hirose T, Terao T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.) Planta. 2004;220:9–16. doi: 10.1007/s00425-004-1314-6. [DOI] [PubMed] [Google Scholar]
- Hirose T, Imaizumi N, Scofield GN, Furbank RT, Ohsugi R. cDNA cloning and tissue specific expression of a gene for sucrose transporter from rice (Oryza sativa L.) Plant Cell Physiol. 1997;38:1389–1396. doi: 10.1093/oxfordjournals.pcp.a029134. [DOI] [PubMed] [Google Scholar]
- Hirose T, Takano M, Terao T. Cell wall invertase in developing rice caryopsis: molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol. 2002;43:452–459. doi: 10.1093/pcp/pcf055. [DOI] [PubMed] [Google Scholar]
- Hirose T, Scofield GN, Terao T. An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 2008;174:534–543. doi: 10.1016/j.plantsci.2008.02.009. [DOI] [Google Scholar]
- Hoshikawa K. Studies on the development of endosperm in rice: 1. Process of endosperm tissue formation. Proc Crop Sci Soc Jpn. 1967;36:151–161. doi: 10.1626/jcs.36.151. [DOI] [Google Scholar]
- Hoshikawa K. Studies on the development of endosperm in rice: 11. Development of amyloplasts in endosperm tissue. Proc Crop Sci Soc Jpn. 1968;37:207–216. doi: 10.1626/jcs.37.207. [DOI] [Google Scholar]
- Ishimaru T, Matsuda T, Ohsugi R, Yamagishi T. Morphological development of rice caryopses located at the different positions in a panicle from early to middle stage of grain filling. Funct Plant Biol. 2003;30:1139–1149. doi: 10.1071/FP03122. [DOI] [PubMed] [Google Scholar]
- Ishimaru T, Hirose T, Matsuda T, Goto A, Takahashi K, Sasaki H, Terao T, Ishii R, Ohsugi R, Yamagishi T. Expression patterns of genes encoding carbohydrate-metabolizing enzymes and their relationship to grain filling in rice (Oryza sativa L.): comparison of caryopses located at different positions in a panicle. Plant Cell Physiol. 2005;46:620–628. doi: 10.1093/pcp/pci066. [DOI] [PubMed] [Google Scholar]
- Ishimaru T, Nakazono M, Masumura T, Abiko M, San-oh Y, Nishizawa NK, Kondo M. A method for obtaining high integrity RNA from developing aleurone cells and starchy endosperm in rice (Oryza sativa L.) by laser microdissection. Plant Sci. 2007;173:321–326. doi: 10.1016/j.plantsci.2007.06.004. [DOI] [Google Scholar]
- Ishimaru T, Horigane AK, Ida M, Iwasawa N, San-oh YA, Nakazono M, Nishizawa NK, Masumura T, Kondo M, Yoshida M. Formation of grain chalkiness and changes in water distribution in developing rice caryopses grown under high-temperature stress. J Cereal Sci. 2009;50:166–174. doi: 10.1016/j.jcs.2009.04.011. [DOI] [Google Scholar]
- Itani T, Tamaki M, Arai E, Horino T. Distribution of amylase, nitrogen, and minerals in rice kernels with various characters. J Agri Food Chem. 2002;50:5326–5332. doi: 10.1021/jf020073x. [DOI] [PubMed] [Google Scholar]
- Itoh K, Ozaki H, Okada K, Hori H, Takeda Y, Mitsui T. Introduction of Wx transgenic into rice wx mutants leads to both high- and low-amylose in rice. Plant Cell Physiol. 2003;44:473–480. doi: 10.1093/pcp/pcg068. [DOI] [PubMed] [Google Scholar]
- Kim BR, Nam HY, Kim SU, Kim SI, Chang YJ. Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotech Let. 2003;25:1869–1872. doi: 10.1023/A:1026298032009. [DOI] [PubMed] [Google Scholar]
- Koh HJ, Son HY, Heu MH, Lee HS, McCouch SR. Molecular mapping of a new genic male-sterility gene causing chalky endosperm in rice (Oryza sativa L.) Euphytica. 1999;106:57–62. doi: 10.1023/A:1003575016035. [DOI] [Google Scholar]
- Koyama H, Kihara T, Takita E, Hara T. Isolation of Two cDNA Clones Encoding NADP-Specific Isocitrate Dehydrogenase from Rice. Plant Physiol. 1999;120:1207. [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y. The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 1999;121:399–409. doi: 10.1104/pp.121.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo N, Harada K, Hirai A, Kadowaki K. A single nuclear transcript encoding mitochondria RPS14 and SDHB of rice is processed by alternative splicing: Common use of the same mitochondrial targeting signal for different proteins. PNAS. 1999;96:9207–9211. doi: 10.1073/pnas.96.16.9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Hwang SK, Han M, Eom JS, Kang HG, Han Y, Choi SB, Cho MH, Bhoo SH, An G, Hahn TR, Okita TW, Jeon JS. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.) Plant Mol Biol. 2007;65:531–546. doi: 10.1007/s11103-007-9153-z. [DOI] [PubMed] [Google Scholar]
- Lin CF, Jiang LZ, Zhang XN, Qian XY, Liang ZS, Yang JS. Cloning and prokaryotic expression of a cDNA encoding a putative mitochondrial malate dehydrogenase in Oryza sativa. DNA Seq. 2003;14:75–77. doi: 10.1080/1042517031000062800. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Kimura K, Arai Y, Kawasaki T, Shimada H, Baba T. Starch branching enzymes from immature rice seeds. J Biochem. 1992;112:643–651. doi: 10.1093/oxfordjournals.jbchem.a123953. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Kawasaki T, Shimada H, Satoh H, Kobayashi E, Okumura S, Arai Y, Baba T. Alternation of the structural properties of starch components by the lack of an isoform of starch branching enzyme in rice seeds. J Biol Chem. 1993;268:19084–19091. [PubMed] [Google Scholar]
- Nakamura Y, Umemoto T, Ogata N, Kuboki Y, Yano M, Sasaki T. Starch debranching enzyme (R-enzyme or pullulanase) from developing endosperm: purification, cDNA and chromosomal localization of the gene. Planta. 1996;199:209–218. doi: 10.1007/BF00196561. [DOI] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H. Biochemical and genetic analysis of the effects of Amylose-Extender mutation in rice endosperm. Plant Physiol. 2001;127:459–472. doi: 10.1104/pp.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB, Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot. 2005;56:3229–3244. doi: 10.1093/jxb/eri292. [DOI] [PubMed] [Google Scholar]
- Okagaki RJ. Nucleotide sequence of a long cDNA from the rice waxy gene. Plant Mol Biol. 1992;19:513–516. doi: 10.1007/BF00023402. [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Gates PJ, Boulter D. Regularly aligned mitochondria in aleurone and sub-aleurone layers of developing rice endosperm. Plant Cell Environ. 1981;4:355–357. doi: 10.1111/j.1365-3040.1981.tb02112.x. [DOI] [Google Scholar]
- Patron NJ, Greber B, Fahy BF, Laurie DA, Parker ML, Denyer K. The lys5 mutations of barley reveal the nature and importance of plastidial ADP-Glc transporters for starch synthesis in cereal endosperm. Plant Physiol. 2004;135:2088–2097. doi: 10.1104/pp.104.045203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Xie W, Lian X, Zhang Q. Molecular analyses of the rice glutamate dehydrogenase gene family and their response to nitrogen and phosphorus deprivation. Plant Cell Rep. 2009;28:1115–1126. doi: 10.1007/s00299-009-0709-z. [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Weschke W, Weber H, Wobus U, Borisjuk L. Energy state and its control on seed development: starch accumulation is associated with high ATP and steep oxygen gradients within barley grains. J Exp Bot. 2004;55:1351–1359. doi: 10.1093/jxb/erh130. [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Koch K, Wobus U, Borisjuk L. Positional cues for the starch/lipid balance in maize kernels and resource partitioning to the embryo. Plant J. 2005;42:69–83. doi: 10.1111/j.1365-313X.2005.02352.x. [DOI] [PubMed] [Google Scholar]
- Ryoo N, Yu C, Park CS, Baik MY, Park IM, Cho MH, Bhoo SH, An G, Hahn TR, Jeon JS. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.) Plant Cell Rep. 2007;26:1083–1095. doi: 10.1007/s00299-007-0309-8. [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Ogawa M, Masumura T, Shibata D, Takeba G, Tanaka K, Fujii S. Three cDNA sequences coding for glutamate synthetase polypeptide in Oryza sativa L. Plant Mol Biol. 1989;13:611–614. doi: 10.1007/BF00027323. [DOI] [PubMed] [Google Scholar]
- Satoh H, Shibahara K, Tokunaga T, Nishi A, Tasaki M, Hwang SK, Okita TW, Kaneko N, Fujita N, Yoshida M, Hosaka Y, Sato A, Utsumi Y, Ohdan T, Nakamura Y. Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell. 2008;20:1833–1849. doi: 10.1105/tpc.107.054007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Nishi A, Yamashita K, Takemoto Y, Tanaka Y, Hosaka Y, Sakurai A, Fujita N, Nakamura Y. Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 2003;133:1111–1121. doi: 10.1104/pp.103.021527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield GN, Hirose T, Gaudron JA, Upadhyaya NM, Ohsugi R, Furbank RT. Antisense suppression of the rice sucrose transporter gene, OsSUT1, leads to impaired grain filling and germination but does not affect photosynthesis. Funct Plant Biol. 2002;29:815–826. doi: 10.1071/PP01204. [DOI] [PubMed] [Google Scholar]
- Shannon JC, Pien FM, Cao H, Liu KC. Brittle-1, an adenylate translocator, facilitates transfer of extraplastidial synthesized ADP-glucose into amyloplasts of maize endosperms. Plant Physiol. 1998;117:1235–1252. doi: 10.1104/pp.117.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura S, Yamamoto R, Nakamura T, Shimada S, Komatsu S. Stem hypertrophic lenticels and secondary aerenchyma enable oxygen transport to roots of soybean in flooded soil. Ann Bot. 2010;106:277–284. doi: 10.1093/aob/mcq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikka VK, Choi SB, Kavakli H, Sakulsingharoj C, Gupta S, Ito H, Okita TW. Subcellular compertmentation and allosteric regulation of the rice endosperm ADPglucose pyrophosphorylase. Plant Sci. 2001;161:461–468. doi: 10.1016/S0168-9452(01)00431-9. [DOI] [Google Scholar]
- Sturm A, Tang GQ. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 1999;4:401–407. doi: 10.1016/S1360-1385(99)01470-3. [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Sugiyama K, Ishiyama K, Inoue E, Sato T, Takahashi H, Yamaya T. Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase 1;1. Plant J. 2005;42:641–651. doi: 10.1111/j.1365-313X.2005.02406.x. [DOI] [PubMed] [Google Scholar]
- Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell. 2004;16:694–708. doi: 10.1105/tpc.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota K, Tamura M, Ohdan T, Nakamura Y. Expression profiling of starch metabolism-related plastidic translocator genes in rice. Planta. 2006;223:248–257. doi: 10.1007/s00425-005-0128-5. [DOI] [PubMed] [Google Scholar]
- Umemoto T, Aoki N, Lin HX, Nakamura Y, Inouchi N, Sato Y, Yano M, Hirabayashi H, Maruyama S. Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct Plant Biol. 2004;31:671–684. doi: 10.1071/FP04009. [DOI] [PubMed] [Google Scholar]
- Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, Ma H, Zhang G, He Z. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet. 2008;40:1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- Woo MO, Ham TH, Ji HS, Choi MS, Jiang W, Chu SH, Piao R, Chin JH, Kim JA, Park BS, Seo HS, Jwa NS, McCouch SR, Koh HJ. Inactivation of the UGPase1 gene causes genic male sterility and endosperm chalkiness in rice (Oryza sativa L.) Plant J. 2008;54:190–204. doi: 10.1111/j.1365-313X.2008.03405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SB, Zhu TL, Deng Y, Chong K, Xue Y, Wang T. Dynamic proteomic analysis reveals a switch between central carbon metabolism and alcoholic fermentation in rice filling grains. Plant Physiol. 2008;148:908–925. doi: 10.1104/pp.108.125633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Kubota T, Masumura T, Shiraishi N, Tanaka K, Sugimoto T, Oji Y. Molecular cloning, gene expression and functional expression of a phosphoenolpyruvate carboxylase Osppc1 in developing rice seeds: implication of involvement in nitrogen accumulation. Seed Sci Res. 2014;24:23–36. doi: 10.1017/S0960258513000354. [DOI] [Google Scholar]


