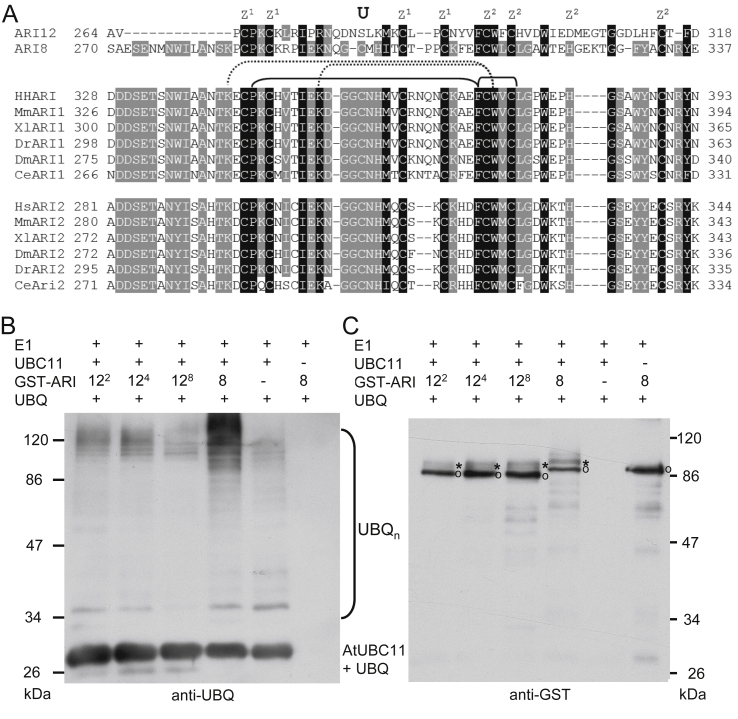
Fig. 1.
Sequence alignment of the RING2 domain and ubiquitination assays of ARI12 and ARI8. (A) Conserved residues between ARI12 and the structurally characterized HHARI and homologs from mouse (Mm), frog (Xl), zebra fish (Dr), fly (Dm) and worm (Ce). Cysteine and histidine residues coordinating the two Zn2+ atoms are marked with Z1 and Z2. The conserved cysteine forming the thiolester bond with glycine of ubiquitin is marked with U. Note that only in ARI12 this residue is serine. Conserved key contacts for F371 and W373 revealed in HHARI are marked with arches. Residues 100% and 90% identical between the sequences are highlighted with black and grey, respectively. (B, C) In vitro ubiquitination assays with GST-tagged full-length ARI12 and ARI8 mediate polyubiquitination. Omission of AtUBC11 and of GST-ARI8 resulted in a loss of protein polyubiquitination. Ubiquitinated proteins were visualized via Western blot analysis using ubiquitin (UBQ) (B) or anti-GST antibodies. Position and size of molecular markers are on the side of each blot. * mark the monoubiquitinated GST-tagged ARI proteins and open circle the GST-tagged ARI and UBC11 proteins. Superscript numbers indicated the increasing amount of GST-tagged ARI12 used in the assays.