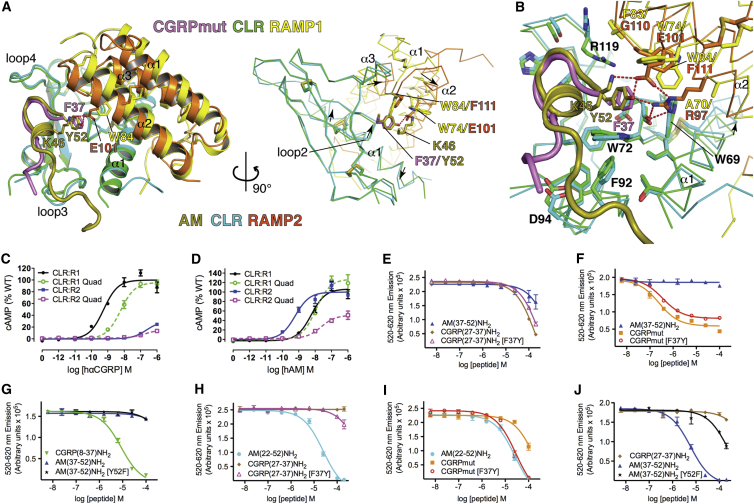
Figure 6.
Peptide Selectivity Determinants for CLR:RAMP1/2 Complexes
(A) Superposition of the CGRPmut- and AM-bound ECD heterodimers aligned based on the CLR positions. The receptors and peptides are in cartoon representation with selected residues as sticks in the left image. In the right image, the receptors are Cα traces and the peptide cartoons were omitted for clarity. Arrows indicate directions of movement of CLR α1 and loop 2 and RAMP α2 and α3 from the CGRPmut/RAMP1-bound state to the AM/RAMP2-bound state. Red dashes are hydrogen bonds.
(B) Detailed view of the aligned CGRPmut- and AM-bound complexes with selected residues as sticks and the receptors as Cα traces. The arrow highlights the shift of the C-terminal region of CLR α1 from the CGRPmut/RAMP1- to AM/RAMP2-bound states.
(C and D) Concentration-response curves for the CGRP and AM1 receptors with the RAMP1 (A70R, W74E, F83G, W84F) and RAMP2 (R97A, E101W, G110F, F111W) quadruple “swap” mutants tested with hαCGRP and hAM in cAMP assays in COS-7 cells. The cell surface expression was: RAMP1 quad 116.4 ± 5.25 (n = 4) % WT, RAMP2 quad 73.7 ± 9.52 (n = 4) % WT p < 0.01 by one-way ANOVA followed by Dunnett’s test for the RAMP2 quad mutant.
(E–J) Competition AlphaScreen assays with purified receptor ECD heterodimer proteins and the indicated competitor “swap” peptides. (E)–(G) are for the MBP-RAMP1-(GSA)3-CLR-H6 protein with biotin-CGRP (100 nM each) and (H)–(J) are for the MBP-RAMP2[L106R]-(GSA)3-CLR-H6 protein with biotin-AM (100 nM each). The binding data are representative of at least three independent experiments each performed in duplicate. The error bars represent the SEM of the experiment. Determinable pIC50 values were as follows: (F), CGRPmut 6.79 ± 0.10 and CGRPmut [F37Y] 6.24 ± 0.09; (G), CGRP(8-37) 5.01 ± 0.04; (H), AM(22-52) 4.92 ± 0.16; (I), AM(22-52) 4.84 ± 0.09 and CGRPmut [F37Y] 4.79 ± 0.07; (J), AM(37-52) 5.00 ± 0.11.