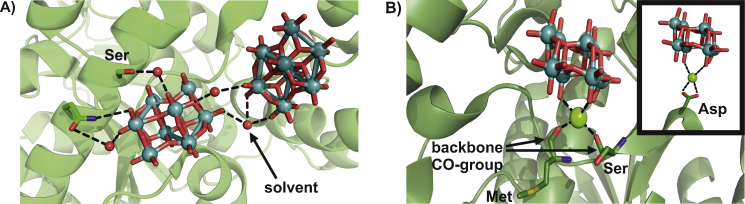
Fig. 3.
Solvent-mediated interactions in Azotobacter vinelandii (PDB entry: 4F6T). (A) Glutamine (Gln) residue interacts directly with a terminal oxygen of an octamolybdate via its Nɛ2 nitrogen atom, whereas the interaction between its Oɛ2 oxygen atom and the octamolybdate is mediated via the solvent (only the oxygen atom of the water is shown as a red sphere). A serine residue is also connected to the same octamolybdate via a solvent molecule. In addition to solvent-mediated POM–protein interactions, the figure also shows a solvent-mediated POM–POM interactions between two neighboring octamolybdates. The hydrogen bond distances between the solvent molecules and the binding partners vary from 2.5 to 3.0 Å (only the distance between the glutamine carbonyl group and the water is greater with 3.8 Å). (B) The interaction between two protein backbone carbonyl groups and the terminal oxygen atoms of an octamolybdate is mediated by a Mg2+ ion. The distances between the Mg2+ ion and the binding partners are about 2.4–2.5 Å. The inset in the same figure shows the theoretical possible interaction of a negatively charged octamolybdate with a negatively charged side chain (aspartic acid) mediated by a Mg2+ ion. The protein is depicted as a cartoon (30–50% transparency) with interacting side chains shown as ball and stick (color code: carbon atoms = green/dark green, nitrogen atom = blue and oxygen atoms = red, sulfur atom = yellow) and the magnesium ion as light green sphere. The octamolybdates are illustrated in ball and stick (color code: molybdenum atoms = deep teal, oxygen atoms = red).