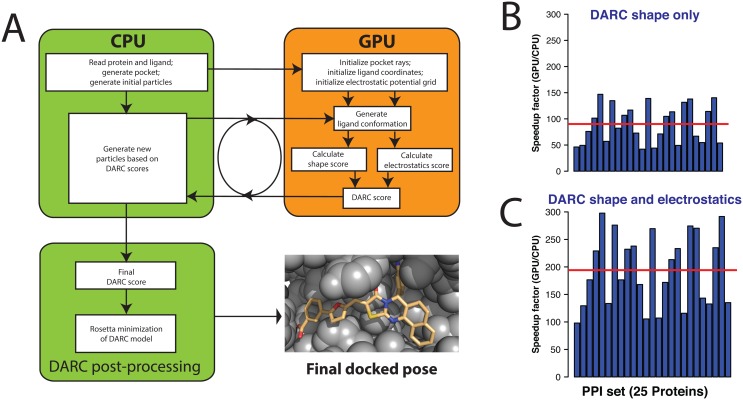
Fig 6. Updated GPU control flow.
(A) Schematic illustration of CPU-GPU control flow in DARC 2.0. Previously, the ligand conformation was generated on the CPU and passed to the GPU; now, the conformer index / displacement / rotation (relative to a “reference” position) is instead passed, and the GPU is responsible for applying this transformation to the ligand’s atomic coordinates. The new electrostatic complementarity term is also computed entirely on the GPU. (B) For each protein-ligand complex in our set (S1 Table), we timed DARC when docking the ligand back into its cognate receptor, using either GPU+CPU or CPU alone. We find an average speedup of 90-fold when using the GPU (red line), an improvement over the 27-fold speedup we achieved in our original GPU implementation of DARC [26]. (C) The GPU led to an even greater speedup over the analogous calculation on the CPU when electrostatic complementarity was included in both calculations (190-fold speedup).