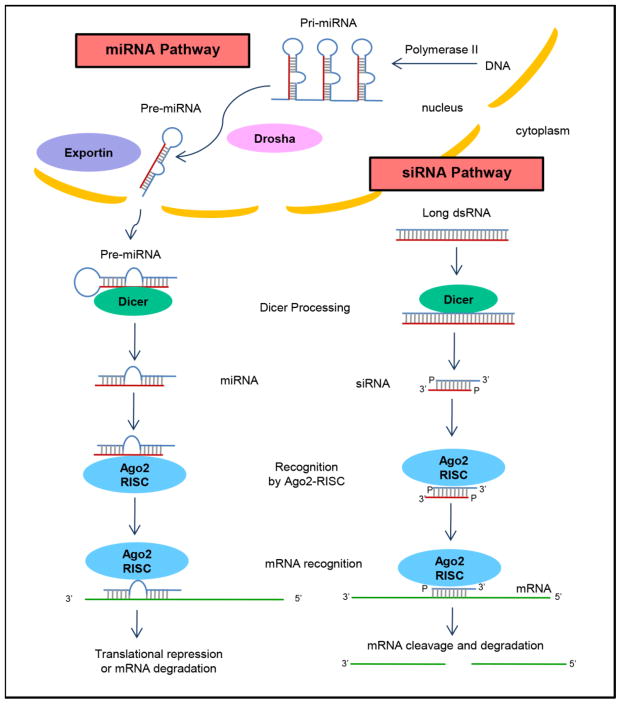
Fig. 1.
The process of RNA interference in eukaryotic cells. Long precursor miRNA (called pri-miRNA) is processed by Drosha into pre-miRNAs in the nucleus. Following transportation to the cytoplasm via exportin-5, pre-miRNAs are further processed by an RNAse III enzyme called Dicer to produce shorter double-stranded miRNAs with imperfect complementarity. miRNAs are recognized by Argonaute 2 (AGO2) and RNA-induced silencing complex (RISC), where one of the strands is degraded and the other strand guides the AGO2-RISC complex to bind and block translation of target mRNAs having partial complementary sites. Cytoplasmic long double-stranded RNA (dsRNA) is cleaved by the cytosolic enzyme Dicer into small interfering RNAs (siRNAs). siRNAs are recognized by the AGO2-RISC enzyme complex. One of the strands is degraded and the other strand guides the complex to recognize mRNA sequences with perfect or nearly perfect complementarity, resulting in cleavage and degradation of target.